Degradation of deproteinized natural rubber by Gordonia sp. isolated from enrichment consortia
Biodegradation is a potential way of decomposing deproteinized natural rubber
(DPNR). The enrichment consortia were demonstrated from a rubber processing factory waste.
Nine DPNR-degrading bacteria were isolated from those consortia. The highest DPNR film
weight loss in a mineral salt medium (MSM) was 43.92 ± 2.30 % after 30 days incubation using
strain 5A1. The formation of aldehyde group during rubber degradation of 5A1 was determined
using Schiff staining and Fourier Transform Infrared spectroscopy (FTIR) analysis. The 16S
rRNA gene sequence, of 5A1 showed the highest identity with that of Gordonia soli CC-AB07.
This is the first report to demonstrate a strong ability to degrade DPNR by Gordonia sp. isolated
from a rubber processing factory waste in Viet Nam

Trang 1

Trang 2

Trang 3

Trang 4
Trang 5

Trang 6

Trang 7

Trang 8
Tóm tắt nội dung tài liệu: Degradation of deproteinized natural rubber by Gordonia sp. isolated from enrichment consortia

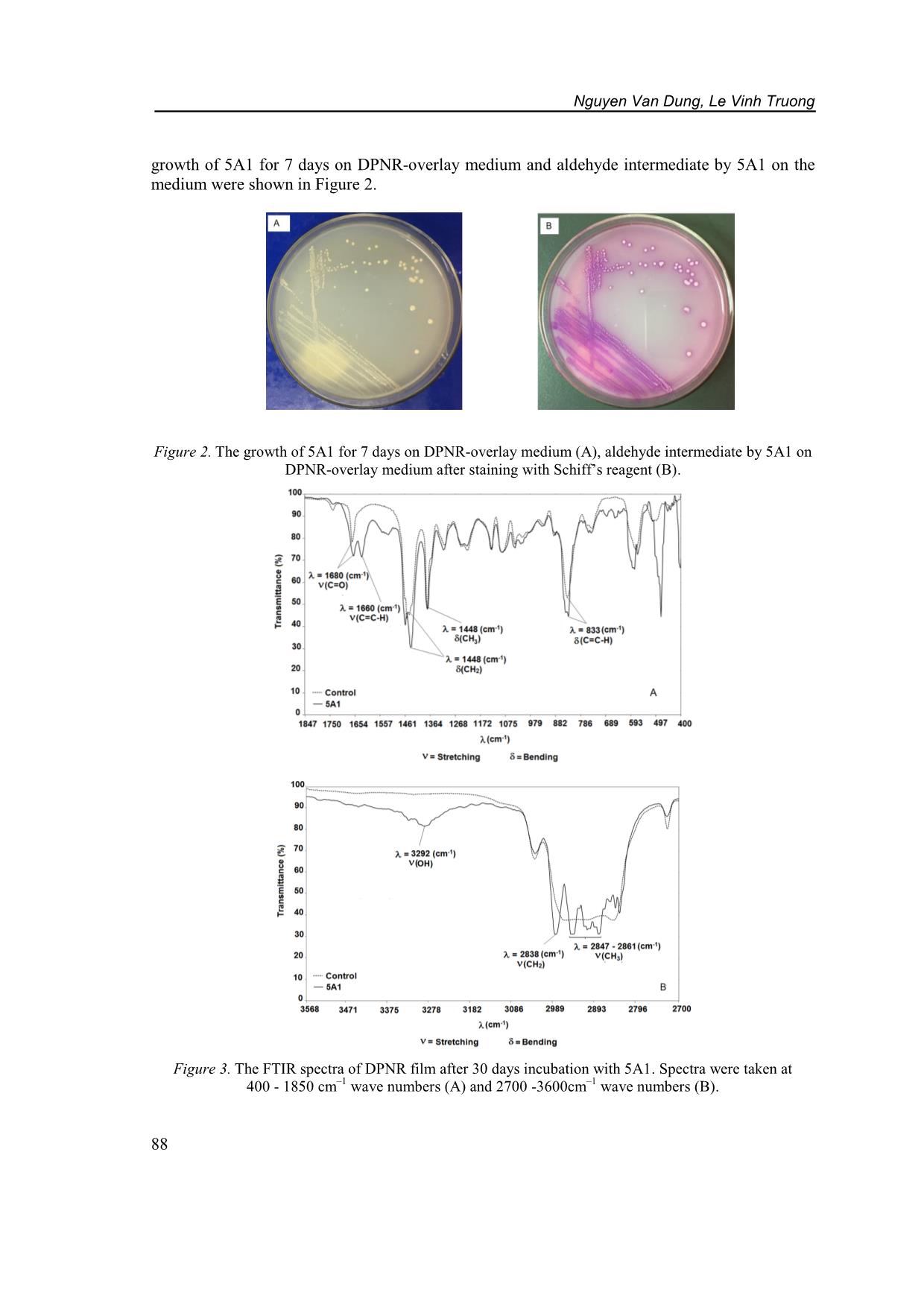
-overlay agar plates for further study. The individual isolated strain was incubated in 20 mL of MSM supplemented with DPNR films (0.3 %, w/v) as a sole carbon source at 30 ◦C, 150 rpm for 30 days. Each experiment was performed in duplicate. After incubation, DPNR films were collected and washed with 0.1 M NaOH for 30 minutes followed by distilled water to remove attached biomass. Samples were then dried at 50 °C until a constant weight [3]. The weight loss of DPNR films was measured before (W1) and after degradation (W2). Weight loss (%) was calculated as follows: Weight loss = x 100 2.2.2. Deproteinized natural rubber degradation by selected isolate The selected strain was obtained based on the highest DPNR film weight loss after 30 days incubation. It was grown on a DPNR-overlay plate after 7 days and stained with Schiff’s reagent. The purple colour produced by the reagent shows the evidence that polyisoprene oligomers containg aldehyde group was accumulated during the microbial degradation of rubber. Staining was performed as described previously [9]. The FTIR (Jasco-4600, Japan) analysis was done to detect the degradation of DPNR film after 30 days inculation by selected strain in MSM at 30 oC and 150 rpm on the basis of changes in the functional group. The DPNR films was mixed with KBr and made into a pellet, which was fixed to the FTIR sample plate. Spectra were taken at 400 to 4000 cm–1 wave numbers for each sample. The correlation of absorption bands in the spectrum of an unknown compound with the known absorption frequencies were analyzed [10]. 2.2.3. 16S RNA gene analysis The total genomic DNA of the selected strain was extracted using a QIAamp DNA mini kit (QIAGEN). The 16S rRNA gene sequences were amplified using the bacterial universal primers named 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5'- GGTTACCTTGTTACGACTT-3') [21]. The PCR reaction was carried out in a final volume of 20 µl containing 10 µl PCR Master mix 2x Promega Taq DNA Polymerase, 1 µl primer 10 µM 27F, 1 µl primer 10 µM 1492R, 1 µl of the DNA template and 7 µl MiliQ. The PCR was performed according to the following program: 2 min denaturation at 94 oC, followed by 30 cycles of 1 min denaturation at 94 oC; 1 min annealing at 50 oC; 1 min extension at 72 oC; and a final extension step of 2 min at 72 oC. The 16S rDNA gene was sequenced. The sequences analysis was carried out on an ABI Prism, 3100-Avant Genetic Analyzer (Hitachi, Japan). The gene sequences were aligned with published sequence in Genbank database using the basic local alignment search tool (BLAST) at the United State National Center of Biotechnology 86 Finite element modelling for electric field distribution around positive streamers in oil Information. A phylogenetic analysis was performed with the Clustal W program using the neighbour joining method. 2.2.4. Nucleotide sequence accession number The partial 16S rRNA nucleotide sequence of the selected strain has been deposited in the GenBank database. 3. RESULTS AND DISCUSSION 3.1. Screening of deproteinized natural rubber degrading bacteria To enhance the growth of deproteinized natural rubber degrading bacteria, the enrichment consortia were demonstrated with five sub-transfers. The nine isolates obtained from enrichment consortia were able to grow on MSM agar containing DPNR as the sole carbon source. Only one of isolate, 1A2 formed a clearing zone around the colony and other isolates did not show the clearing zone around the colonies. However, all isolates grew well in the liquid MSM with DPNR films. The degradation of rubber by isolates was determined based on the DPNR film weight loss in Figure 1. Figure 1. DPNR film weight loss after 30 days incubation with isolates. After 30 days incubation, the weight loss of DPNR film by isolates varied from 9.31 ± 1.32 to 43.92 ± 2.30 %. The highest weight loss was obtained from the culture of strain 5A1. Nawong C. et al. isolated Rhodococcus pyridinivorans strain F5 from soil samples in Thailand, the weight loss of latex glove pieces for 30 days of incubation by F5 at 30oC was 9.36 % [10]. In another study Gallert C. showed the weight loss of latex gloves by Streptomyces sp. strain La7 for 32 days of incubation at 30 oC was 13.5 % [11]. Trang N. et al. reported that four NR degrading strains were isolated from waste in Cam Thuy of Viet Nam belonging to Streptomyces sp. The weight loss of NR pieces of strain NMD1, strain M4D1b, strain M4T1c, and strain T37 at 30 oC for 30 days were 4.28 ± 0.32 %, 3.11 ± 0.5 %, 2.05 ± 0.76 % and 2.43 ± 0.34 %, respectively [12]. It seems that strain 5A1 had strong ability to degrade DPNR. 3.2. Deproteinized natural rubber degrading bacteria by strain 5A1 In order to determine the rubber degradation products contain aldehyde groups, the acumulation of aldehydes on DPNR-overlay agar plate was examined by Schiff’s reagent. The 87 Nguyen Van Dung, Le Vinh Truong growth of 5A1 for 7 days on DPNR-overlay medium and aldehyde intermediate by 5A1 on the medium were shown in Figure 2. Figure 2. The growth of 5A1 for 7 days on DPNR-overlay medium (A), aldehyde intermediate by 5A1 on DPNR-overlay medium after staining with Schiff’s reagent (B). Figure 3. The FTIR spectra of DPNR film after 30 days incubation with 5A1. Spectra were taken at 400 - 1850 cm–1 wave numbers (A) and 2700 -3600cm–1 wave numbers (B). 88 Finite element modelling for electric field distribution around positive streamers in oil A purple color remained around the colonies of 5A1 (Fig. 2B), this result suggests that 5A1 degrades rubber via intermediates with aldehyde as indicated by previous studies [6, 8, and 9]. The FTIR spectroscopy was applied to detect the changes in the functional groups of the polymer. The FTIR analysis of the DPNR films after incubation with 5A1 in MSM broth for 30 days showed various spectrum changes compared to the non-biotic control (Figure 3). Characteristic peaks in DPNR films were found at 833 cm-1 representing −C=CH bending. On the other hand, the transmittance area at 1660 cm-1 was attributed to symmetric −C=CH -1 stretching. The peaks at 1448 and 1375 cm were assigned to C-H bending of CH2 and CH3, -1 respectively. In addition, the peaks appeared at 2838 and 2861 cm corresponding to CH2 and CH3 stretching, respectively. The spectrum demonstrated a change in the signal area of CH2 and CH3 bonds in the polyisoprene chain. Moreover, the FTIR spectrum of DPNR films incubation with 5A1 showed very strong absorption peak at 1680 cm−1 indicating the presence of aldehyde and ketone (C=O) groups while samples of the control was not found. This result suggested that the break of functional groups like C=CH bonds to form aldehyde and ketone groups. This correlates with the result from staining with Schiff's staining on DPNR-overlay agar medium after growth of 5A1. Besides, another strong peak at 3292 cm-1 for OH stretching from the hydroxyl group was observed in the FTIR spectrum of the DPNR films after 30 days of incubation. Thus, the carbonyl group in the DPNR film was completely changed to the hydroxyl group due to the microbial treatment. The appearance of aldehyde, ketone, and hydroxyl groups from FTIR analysis indicated that some double bonds can be oxidized. The appearance of the peaks corresponding to aldehyde and ketone groups after incubation was also reported by Nawong C. et al. [10], Hiessl S. et al. [13], Bosco F. et al. [14], Vidya and Growther L. [15], Andler R. et al. [16]. 3.3. Phylogenetic analysis using 16S RNA gene sequence Figure 4. Phylogenetic tree of 16S rRNA gene sequences of 5A1 with the other known rubber degrading bacteria. The tree was constructed by the neighbor-joining method with bootstrap analyses for 1000 replicates and the bar shows 0.1 substitutions per nucleootide position. The 16S rRNA gene sequence of 5A1 was determined, and shown the highest similarity to that of Godonia soli CC-AB07 (99 %). The isolates was named Gordonia sp. strain 5A1 and 89 Nguyen Van Dung, Le Vinh Truong submitted to GenBank under the accession number MN545427. The phylogenetic tree of 16S rRNA gene sequences of 5A1 with the other known rubber degrading bacteria was constructed by the neighbor joining method in Figure. 4. G. polyisoprenivorans strain VH2 was first the genus Gordonia isolated from soil of a rubber tree plantation had been reported mainly because of its ability to degrade natural and synthetic rubber [17] and the genome of VH2 was sequenced and annotated to elucidate the degradation pathway [6]. G. westfalica strain Kb2 was isolated from foul water held inside a deteriorated automobile tire found on a farmer’s field in Germany. Strain Kb2 was able to solubilize and mineralize natural rubber substrates and synthetic cis-1,4-polyisoprene [5]. G. paraffinivorans MTZ041 isolated from a compost, which was observed as the formation of biofilm-like structures on natural and synthetic rubber [8]. The 16S rRNA gene sequence of strain 5A1, consisting of 1405 nucleotides, was compared to the member of the genus Gordonia. It showed similarity 97 % with the 16S rDNA sequences of G. polyisoprenivorans strain VH2, G. westfalica strain Kb2, and G. paraffinivorans MTZ041. 4. CONCLUSIONS In the present study, the enrichment consortia showed the potential to enhance the growth of deproteinized natural rubber degrading bacteria. Nine DPNR degrading bacteria were isolated. Gordonia sp. strain 5A1 demonstrated the highest rubber degrading activity among all isolates. The formation of aldehyde groups during degradation of DPNR by 5A1 was observed using Schiff staining and FTIR analysis. Further studies are necessary to elucidate the Gordonia sp. 5A1 with other isolates and the bacterial community in consortium. The synergistic interaction of microorganisms will create an alternative approach to perform rubber degradation. Acknowledgement. This research is supported by the Vietnam National Foundation for Science and Technology (NAFOSTED) under grant number 106.04-2017.31. REFERENCES 1. Mooibroek H., and Cornish K. - Alternative sources of natural rubber. Appl. Microbiol. Biotechnol. 53 (2000) 355–365. 2. Huber M.A., and Terezhalmy G.T.- Adverse reactions to latex products: preventive and therapeutic strategies. J. Contemp Dent Pract. 7 (2006) 97-106. 3. Tsuchii, A., Takeda, K., and Tokiwa, Y. - Colonization and degradation of rubber pieces by Nocardia sp. Biodegradation 7 (1996) 41-48. 4. Linos, A., Steinbüchel, A., Spröer, C., and Kroppenstedt, R. M. - Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from automobile tire, Int. J. Syst. Bacteriol. 49 (1999) 1785-1791. 5. Linos, A., Berekaa, M. M., Steinbüchel, A., Kim, K. K., Spröer, C., and Kroppenstedt, R. M. - Gordonia westfalica sp. nov., a novel rubber-degrading actinomycete, Int. J. Syst. Evol. Microbiol. 52 (2002) 1133-1139. 6. Hiessl S., Schuldes J., Thurmer A., Halbsguth T., Broker D., Angelov A., Liebl W., Daniel R., Steinbuchel A. - Involvement of two latex-clearing proteins during rubber degradation and insights into the subsequent degradation pathway revealed by the genome sequence of Gordonia polyisoprenivorans strain VH2. Appl Environ Microbiol. 78 (8) (2012) 2874–2887. 90 Finite element modelling for electric field distribution around positive streamers in oil 7. Berekaa M. M., Lino A., Reichelt R., Keller U., and Steinbuchel A. - Effect of pretreatment of rubbber material on its biodegradability by various rubber degrading bacteria. FEMS Microbiol. Lett. 184 (2000) 199-206. 8. Braga S. P., Santos, A. P., Paganini T., Barbosa D Epamino G. W. C., Morais C., Martins L. F., Silva A. M., Setubal J. C., Vallim M. A., and Pascon R. C. - First report of cis-1,4- polyisoprene degradation by Gordonia paraffinivorans. Brazilian Journal of Microbiology, 2019, https://doi.org/10.1007/s42770-019-00143-w. 9. Linh D.V., Huong N.L., Tabata M., Imai S., Iijima S., Kasai D., Anh T.K., Fukuda M. - Characterization and functional expression of a rubber degradation gene of a Nocardia degrader from a rubber-processing factory.Journal of Bioscience and Bioengineering 123 (4) (2017) 412-418. 10. Nawong C., Umsakul K., and Sermwittayawong N. - Rubber gloves biodegradation by a consortium, mixed culture and pure culture isolated from soil samples.Brazilian Journal of Microbiology 49(3) (2018) 481-488. 11. Gallert C. - Degradation of Latex and of Natural Rubber by Streptomyces Strain La 7. System. Appl. Microbial. 23 (2000) 433-441. 12. Trang B.T., Linh D.V., Huong N.L., Anh T.K., Nghia P.T., Fukuda M. - Screening of natural rubber-degrading microorganisms from rubber processing factory waste in Viet Nam. Internat. J. Waste Resources 3 (2013) 9-12. 13. Hiessl S, Böse D, Oetermann S, Eggers J, Pietruszka J, Steinbüchel A. - Latex clearing protein an oxygenase cleaving poly(-1,4-isoprene) rubber at the cis double bond. Appl Environ Microbiol. 80 (2014) 5231-5240. 14. Bosco F., Antonioli D., Casale A., Gianotti V., Mollea C., Laus M., Malucelli G. - Biodegradation of unvulcanized natural rubber by microorganisms isolated from soil and rubber surface: A preliminary study. Bioremediation Journal 22 (1-2) (2018) 43-52. 15. Vidya T. V. and Growther L. - Biodegradation of rubber using actinobacteria isolated from rubber contaminated soil, Indian Journal of Environmental Protection 37 (2017) 1016-1025. 16. Andler R., Altenhoff A. L., Masing F., Steinbüchel A. - In vitro studies on the degradation of poly(cis-1,4-isoprene), Biotechnol Prog. 34 (4) (2018) 890-899. 17. Linos A., Steinbüchel A. – Microbial degradation of natural and synthetic rubber by novel bacteria belonging to the genus Gordonia. Kautsch. Gummi Kunstst. 51 (1998) 496-499. 18. Linos A., Berekaa M.M., Reichelt R., Keller U., Schmitt J., Flemming H., Kroppenstedt R.M., and Steinbuchel A. - Biodeg radation of cis-1,4-Polyisoprene Rubbers by Distinct Actinomycetes: Microbial Strategies and Detailed Surface Analysis. Applied and Environmental Microbiology 66 (4) (2000) 1639-1645. 19. Yikmis, M. and Steinbüchel, A. - Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber, Appl. Environ. Microbiol. 78 (2012) 4543-4551. 20. Kawahara, S.; Klinklai, W.; Kuroda, H.; Isono, Y. - Removal of proteins from natural rubber with ure. Polym. Adv. Technol. 15 (4) (2004) 181-184. 21. Lane D.J. - 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M. - Nucleic acid techniques in bacterial systematics, New York: Wiley, 1991, 115–175. 91
File đính kèm:
 degradation_of_deproteinized_natural_rubber_by_gordonia_sp_i.pdf
degradation_of_deproteinized_natural_rubber_by_gordonia_sp_i.pdf

