Phylogenetic and phylogenomic analyses for large datasets
Abstract: The phylogenetic tree is a main tool to study the evolutionary relationships among species. Computational methods for building phylogenetic trees from gene/protein sequences have been developed for decades and come of age. Efficient approaches, including distance-Based methods, maximum likelihood methods, or classical maximum parsimony methods, are now able to analyze datasets with thousands of sequences. The advanced sequencing technologies have resulted in a huge amount of data including whole genomes. A number of methods have been proposed to analyze the wholegenome datasets, however, numerous challenges need to be addressed and solved to translate phylogenomic inferences into practices. In this paper, we will analyze widely-used methods to construct large phylogenetic trees, and available methods to build phylogenomic trees from whole-genome datasets. We will also give recommendations for best practices when performing phylogenetic and phylogenomic analyses. The paper will enable researchers to comprehend the state-ofthe-art methods and available software to efficiently study the evolutionary relationships among species from large datasets
Trang 1

Trang 2
Trang 3
Trang 4
Trang 5
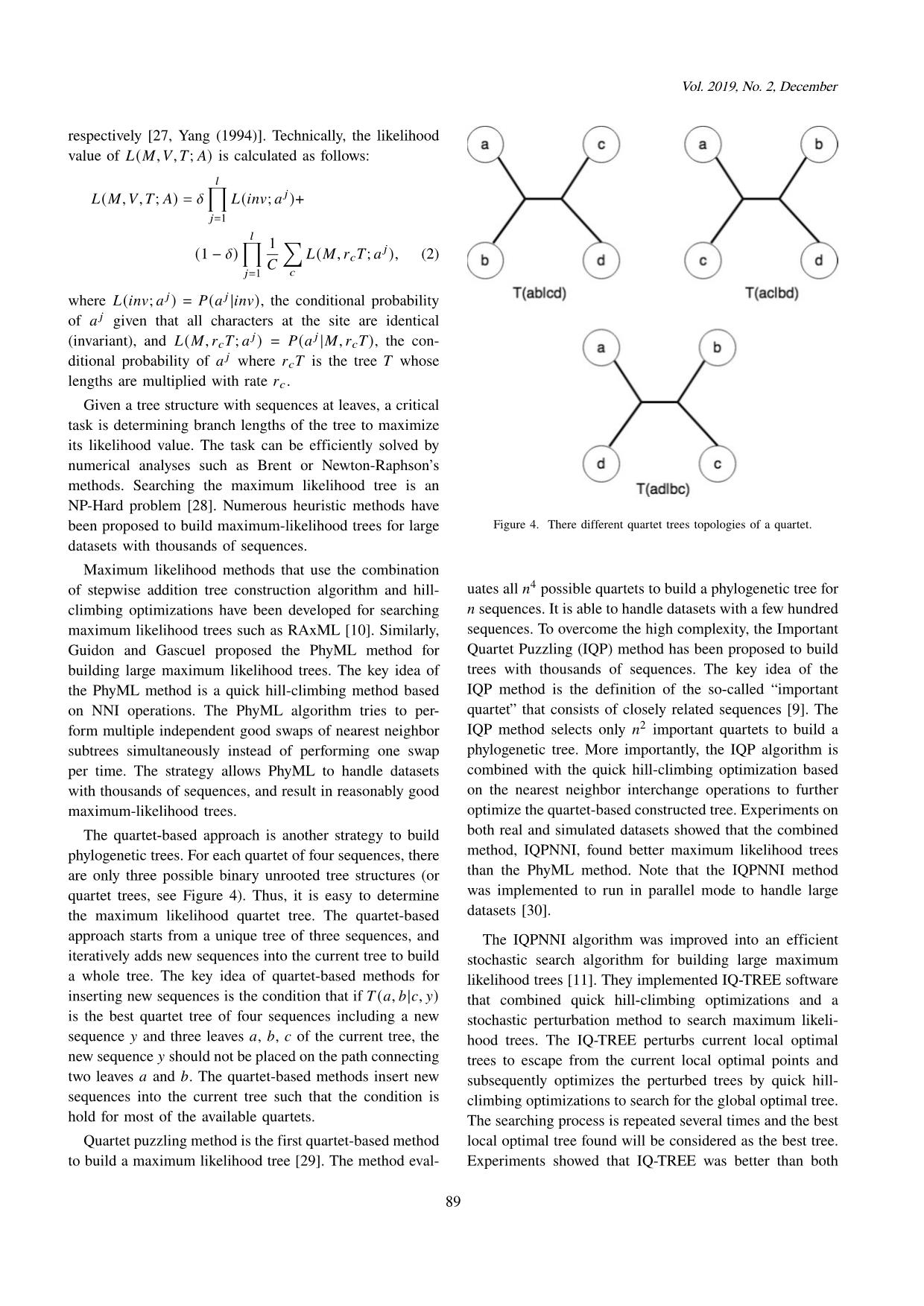
Trang 6

Trang 7

Trang 8

Trang 9
Tóm tắt nội dung tài liệu: Phylogenetic and phylogenomic analyses for large datasets
an efficient iteratively adds new sequences into the current tree to build stochastic search algorithm for building large maximum a whole tree. The key idea of quartet-based methods for likelihood trees [11]. They implemented IQ-TREE software inserting new sequences is the condition that if ) (0, 1|2, H) that combined quick hill-climbing optimizations and a is the best quartet tree of four sequences including a new stochastic perturbation method to search maximum likeli- sequence H and three leaves 0, 1, 2 of the current tree, the hood trees. The IQ-TREE perturbs current local optimal new sequence H should not be placed on the path connecting trees to escape from the current local optimal points and two leaves 0 and 1. The quartet-based methods insert new subsequently optimizes the perturbed trees by quick hill- sequences into the current tree such that the condition is climbing optimizations to search for the global optimal tree. hold for most of the available quartets. The searching process is repeated several times and the best Quartet puzzling method is the first quartet-based method local optimal tree found will be considered as the best tree. to build a maximum likelihood tree [29]. The method eval- Experiments showed that IQ-TREE was better than both 89 Research and Development on Information and Communication Technology TABLE III The second challenge when analyzing the genomes is THE STRUCTURAL VARIANTS IN THE GENOMES. model selection. The evolutionary models including rate 1 2 3 4 5 6 7 8 9 models and substitution models might vary among loci. One Human G1 G2 G3 G4 G5 G6 G7 G8 G9 model cannot properly reflex the evolutionary process of all Gorilla G1 G7 G6 G5 G4 G3 G7 G8 G9 loci. The phylogenomic analyses should use model selection Monkey - G5 G6 G7 G8 G9 G2 G3 G4 methods to assign a proper model for each locus. As esti- Dog - G5 G6 G7 G8 G9 G4 G3 G2 mating model parameters is not strongly affected by the tree structure, model selection methods normally include two steps: building an initial tree and estimating model param- PhyML and RAxML methods in a majority number of cases eters based on the initial tree and the alignment. Building tested. The IQ-TREE software is user-friendly and widely an initial tree can be done efficiently by distance-based used by biologists for studying the evolution of species from methods such as NJ method. Provided that the initial tree is molecular data. reasonably close to the best tree, it is good enough to esti- mate model parameters. Estimating model parameters for an IV. PHYLOGENOMIC ANALYSIS alignment based on the initial tree can be solved by numeri- cal optimization methods such as Brent’s algorithm or more Analyzing whole genomes to investigate the relationships efficient Broyden–Fletcher–Goldfarb–Shanno (BFGS) al- among species is a comprehensive and challenging prob- gorithm. Software like IQ-TREE provides us options to lem. In phylogenetic analysis, a genome is separated into automatically determine the best model for each locus when a list of loci each corresponds to a gene or a region of analyzing whole-genome datasets. Note that normally we interest in the genome. Homologous loci are aligned to do not optimize parameters of amino acid models from create multiple sequence alignments. As a result, the input each alignment because each alignment does not contain for phylogenomic analysis is a list of multiple sequence sufficient data to estimate a large number of parameters. alignments. Phylogenetic tree construction approaches have Finally, the computational expense is a critical burden been expanded to build phylogenomic trees from a list of of phylogenomic inferences. A genome dataset contains multiple sequence alignments. several dozens to thousands of genomes with the length The first challenge in analyzing whole genomes is the up to several hundred million nucleotides. To overcome occurrence of structural variants in the genomes. Besides the problem, more efficient heuristic algorithms in terms variants occurring inside genes, other common types of of both running time and memory requirement should be structural changes are gene insertions/deletions, gene in- developed to handle whole-genome datasets. As genomes versions (inverting the order of genes in the genome), are separated into loci, parallel computing is a promising gene transpositions (moving a number of genes in the approach to perform phylogenomic inference on individual genome from one position to another position in the alignments simultaneously. Most of the current widely-used genome), and inverted transpositions (combining both gene phylogenetic software such as RAxML or IQ-TREE provide inversion and gene transposition events into one event). options to conduct phylogenetic inferences in parallel. Table III demonstrates an example of structural variants in the genomes, e.g., there is a gene inversion between the V. DISCUSSIONS human genome and gorilla genome (genes G3, G4, G5, G6 in the human genome were inverted into G6, G5, G4, G3 Phylogenetic inference is a core study in molecular biol- in the gorilla genome). ogy. It is an active research field for several decades and the The structural changes resulted in genomes with different main focus of prominent researcher groups. Phylogenetic structures. The structural difference between genomes can reconstruction for single or several genes perhaps come to be used as phylogenetic signals for studying the evolution age. The distance-based methods are able to build large rea- of species, i.e., the number of structural changes to explain sonable phylogenies that could be used as starting points to the structural difference between two genomes can be search maximum parsimony or maximum likelihood trees. considered as the genetic distance between the genomes Nowadays, maximum likelihood methods such as RAxML, to evaluate their relationships. Overall, the genetic distance PhyML or IQ-TREE are able to efficiently construct trees between two genomes is a combination of character changes with thousands of sequences. inside genes and structural changes. Weighting and com- All popular phylogenetic tree reconstruction software are bining these changes properly for estimating the overall based on heuristic search methods, therefore, the results genetic distance between genomes enable us to build better from different software, or even from different runs of phylogenetic trees using distance-based methods [31]. the same software, might not completely congruence. It is 90 Vol. 2019, No. 2, December especially true when analyzing datasets whose phylogenetic [7] A. Varón, L. S. Vinh, and W. C. Wheeler, “POY version 4: signals support polytomy tree structures. We recommend phylogenetic analysis using dynamic homologies,” Cladis- tics, vol. 26, no. 1, pp. 72–85, 2010. researchers to perform bootstrap analyses to assess the [8] S. Guindon and O. Gascuel, “A simple, fast, and accurate reliability of branches in the constructed tree. Although algorithm to estimate large phylogenies by maximum like- phylogenetic bootstrapping is computationally expensive, lihood,” Systematic Biology, vol. 52, no. 5, pp. 696–704, ultrafast bootstrap methods such as UFBoot2 [32] are able 2003. [9] L. S. Vinh and A. von Haeseler, “IQPNNI: moving fast to build large bootstrap trees in an acceptable time. through tree space and stopping in time,” Molecular Biology and Evolution, vol. 21, no. 8, pp. 1565–1571, 2004. Determining proper evolutionary models (i.e., site rate [10] A. Stamatakis, “Using RAxML to infer phylogenies,” Cur- models and/or substitution models) for datasets under the rent Protocols in Bioinformatics, vol. 51, no. 1, pp. 6–14, study is very critical in phylogenetic inferences. Using 2015. wrong evolutionary models in analyzing data will lead to [11] L.-T. Nguyen, H. A. Schmidt, A. Von Haeseler, and B. Q. Minh, “IQ-TREE: a fast and effective stochastic algorithm inaccurate results [32]. The evolutionary models are nor- for estimating maximum-likelihood phylogenies,” Molecular mally selected from a list of existing models, and the model Biology and Evolution, vol. 32, no. 1, pp. 268–274, 2015. parameters can be directly estimated from the input data. [12] J. Thompson, D. Higgins, and T. Gibson, “ClustalW,” Nu- cleic Acids Research, vol. 22, no. 22, pp. 4673–4680, 1994. The advance of sequencing technologies has produced [13] R. C. Edgar, “MUSCLE: multiple sequence alignment with large genome datasets consisting of dozens to thousands of high accuracy and high throughput,” Nucleic Acids Research, vol. 32, no. 5, pp. 1792–1797, 2004. genomes with the length up to billion nucleotides. The large [14] F. Joseph, Inferring Phytogenies. Sunderland, MA, USA: genome datasets provide us an unprecedented opportunity Sinauer Associates, 2003. to study the relationships among species from their whole [15] Z. Yang, “Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over genomes. However, new efficient computational methods sites.” Molecular Biology and Evolution, vol. 10, no. 6, pp. should be developed for phylogenomic inferences. The 1396–1401, 1993. relationships among genomes should be analyzed at both [16] S. Whelan and N. Goldman, “A general empirical model levels: point level (i.e., nucleotide/amino acid changes) and of protein evolution derived from multiple protein families using a maximum-likelihood approach,” Molecular Biology structural level (gene insertions/deletions as well as gene re- and Evolution, vol. 18, no. 5, pp. 691–699, 2001. arrangements). Combining changes at both levels to have a [17] S. Q. Le and O. Gascuel, “An improved general amino comprehensive evaluation is a new challenge for researchers acid replacement matrix,” Molecular Biology and Evolution, vol. 25, no. 7, pp. 1307–1320, 2008. in phylogenomic analyses. Another challenge in analyzing [18] C. C. Dang, Q. S. Le, O. Gascuel, and V. S. Le, “FLU, an whole genomes is the heterogeneity of evolutionary pro- amino acid substitution model for influenza proteins,” BMC cesses between loci. Thus, determining proper evolutionary Evolutionary Biology, vol. 10, no. 1, p. 99, 2010. [19] D. T. Jones, W. R. Taylor, and J. M. Thornton, “The models is very critical when analyzing multiple genes or rapid generation of mutation data matrices from protein whole genomes. Currently, several software such as IQ- sequences,” Computational Applied Bioinformatics, vol. 8, TREE are able to perform phylogenomic inferences for large no. 3, pp. 275–282, 1992. genome datasets. [20] L. L. Cavalli-Sforza and A. W. Edwards, “Phylogenetic anal- ysis: models and estimation procedures,” American Journal of Human Genetics, vol. 19, pp. 233–257, 1967. [21] A. Rzhetsky and M. Nei, “Theoretical foundation of REFERENCES the minimum-evolution method of phylogenetic inference.” Molecular Biology and Evolution, vol. 10, no. 5, pp. 1073– [1] N. Saitou and M. Nei, “The neighbor-joining method: a new 1095, 1993. method for reconstructing phylogenetic trees.” Molecular [22] W. H. Day and D. Sankoff, “Computational complexity of Biology and Evolution, vol. 4, no. 4, pp. 406–425, 1987. inferring phylogenies by compatibility,” Systematic Biology, [2] O. Gascuel, “BIONJ: an improved version of the NJ algo- vol. 35, no. 2, pp. 224–229, 1986. rithm based on a simple model of sequence data.” Molecular [23] O. Gascuel, “BIONJ: an improved version of the NJ algo- Biology and Evolution, vol. 14, no. 7, pp. 685–695, 1997. rithm based on a simple model of sequence data.” Molecular [3] L. S. Vinh and A. von Haeseler, “Shortest triplet clustering: Biology and Evolution, vol. 14, no. 7, pp. 685–695, 1997. reconstructing large phylogenies using representative sets,” [24] A. W. Edwards and Cavalli-Sforza, “The Reconstruction of BMC Bioinformatics, vol. 6, no. 1, p. 92, 2005. Evolution,” Annals of Human Genetics, vol. 27, pp. 105–106, [4] J. C. Wilgenbusch and D. Swofford, “Inferring evolutionary 1963. trees with PAUP*,” Current Protocols in Bioinformatics, [25] W. M. Fitch, “Toward defining the course of evolution: no. 1, pp. 6–4, 2003. minimum change for a specific tree topology,” Systematic [5] P. A. Goloboff, J. S. Farris, and K. C. Nixon, “TNT, a free Biology, vol. 20, no. 4, pp. 406–416, 1971. program for phylogenetic analysis,” Cladistics, vol. 24, no. 5, [26] R. Graham and L. Foulds, “Unlikelihood that minimal phy- pp. 774–786, 2008. logenies for a realistic biological study can be constructed in [6] D. T. Hoang, L. S. Vinh, T. Flouri, A. Stamatakis, A. von reasonable computational time,” Mathematical Biosciences, Haeseler, and B. Q. Minh, “MPBoot: fast phylogenetic maxi- vol. 60, no. 2, pp. 133–142, 1982. mum parsimony tree inference and bootstrap approximation,” [27] Z. Yang, “Maximum likelihood phylogenetic estimation from BMC Evolutionary Biology, vol. 18, no. 1, p. 11, 2018. DNA sequences with variable rates over sites: approximate 91 Research and Development on Information and Communication Technology methods,” Journal of Molecular Evolution, vol. 39, no. 3, Le Sy Vinh obtained PhD in Bioinformat- pp. 306–314, 1994. ics from Heinrich Heine University, Dues- [28] B. Chor and T. Tuller, “Maximum likelihood of evolution- seldorf, Germany 2005, subsequently fol- ary trees is hard,” in Annual International Conference on lowed a postdoc fellowship at American Research in Computational Molecular Biology, 2005, pp. 296–310. Museum of Natural History, NYC from [29] K. Strimmer and A. Von Haeseler, “Quartet puzzling: a 2005 to 2008. He is currently the Dean quartet maximum-likelihood method for reconstructing tree of the Faculty of Information Technology, topologies,” Molecular Biology and Evolution, vol. 13, no. 7, University of Engineering and Technology, pp. 964–969, 1996. Vietnam National University, Hanoi. [30] B. Q. Minh, L. S. Vinh, A. Von Haeseler, and H. A. Schmidt, “PIQPNNI: parallel reconstruction of large maximum likeli- Le Sy Vinh is an expert in phylogenetic analysis, the author hood phylogenies,” Bioinformatics, vol. 21, no. 19, pp. 3794– of widely-used software such as IQPNNI, POY4, UFBoot2. He 3796, 2005. is the group leader of many human genome projects in Viet- [31] L. S. Vinh, A. Varón, and W. C. Wheeler, “Pairwise align- nam including the first Vietnamese human genome, building the ment with rearrangements,” Genome Informatics, vol. 17, comprehensive Vietnamese human genome database, or Autism no. 2, pp. 141–151, 2006. spectrum disorder in Vietnamese children. [32] D. T. Hoang, O. Chernomor, A. Von Haeseler, B. Q. Minh, and L. S. Vinh, “UFBoot2: improving the ultrafast bootstrap approximation,” Molecular Biology and Evolution, vol. 35, no. 2, pp. 518–522, 2018. 92
File đính kèm:
 phylogenetic_and_phylogenomic_analyses_for_large_datasets.pdf
phylogenetic_and_phylogenomic_analyses_for_large_datasets.pdf

