Ant colony optimization based founder sequence reconstruction
Reconstruction of a set of genetic sequences (founders) that can combine together to form given genetic
sequences (e.g. DNA) of individuals of a population is an important problem in evolutionary biology. Such
reconstruction can be modeled as a combinatorial optimization problem, in which we have to find a set of
founders upon that genetic sequences of the population can be generated using a smallest number of
recombinations. In this paper we propose an ant colony optimization algorithm (ACO) based method, equipped
with some important improvements, for the founder DNA sequence reconstruction problem. The proposed
method yields excellent performance when validating on 108 test sets from three benchmark datasets. Comparing
with the best by far corresponding method, our proposed method performs better in 45 test sets, equally well in
44 and worse only in 19 sets. These experimental results demonstrate the efficacy and perspective of our
proposed method.

Trang 1

Trang 2

Trang 3
Trang 4

Trang 5
Trang 6

Trang 7
Tóm tắt nội dung tài liệu: Ant colony optimization based founder sequence reconstruction

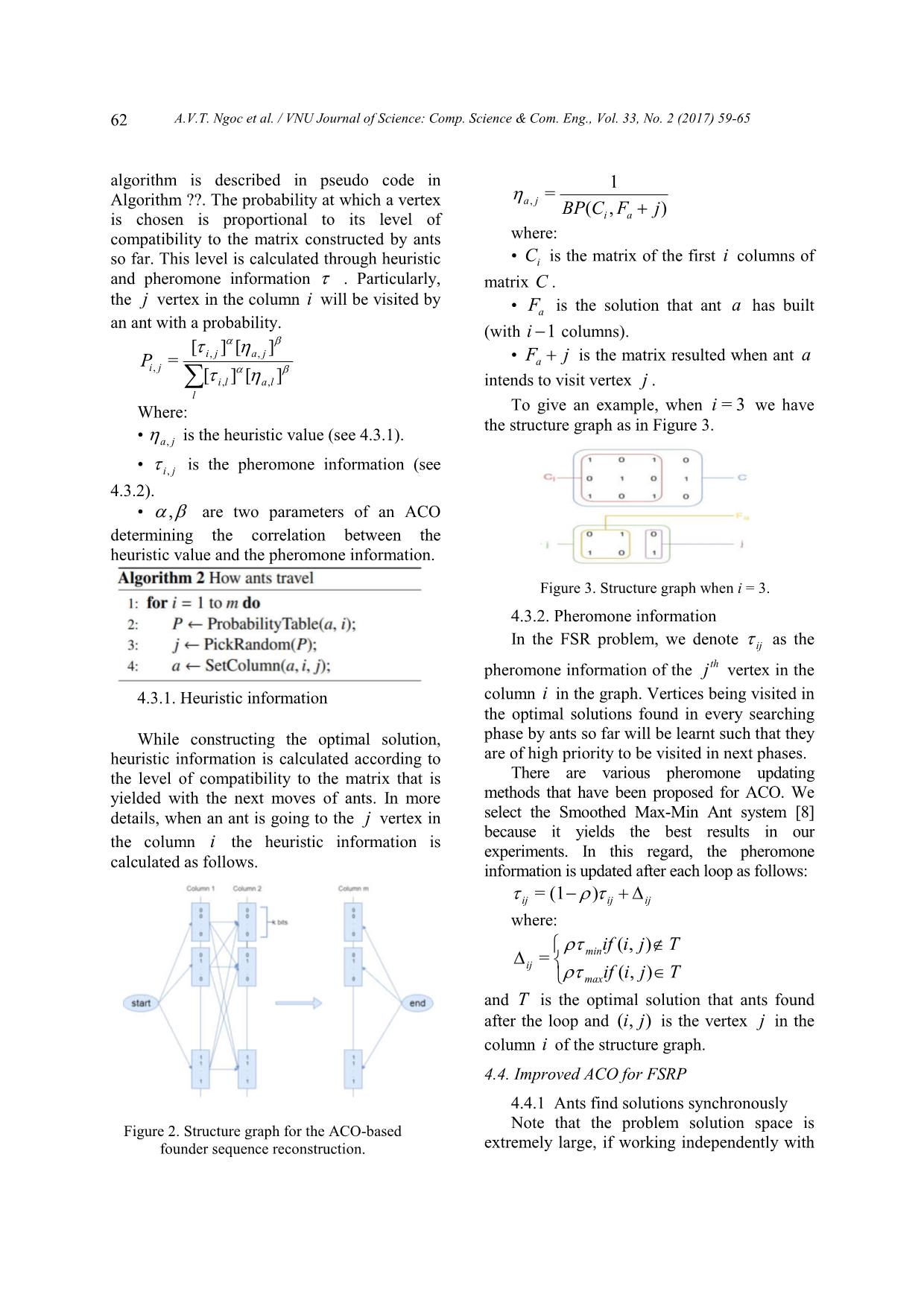
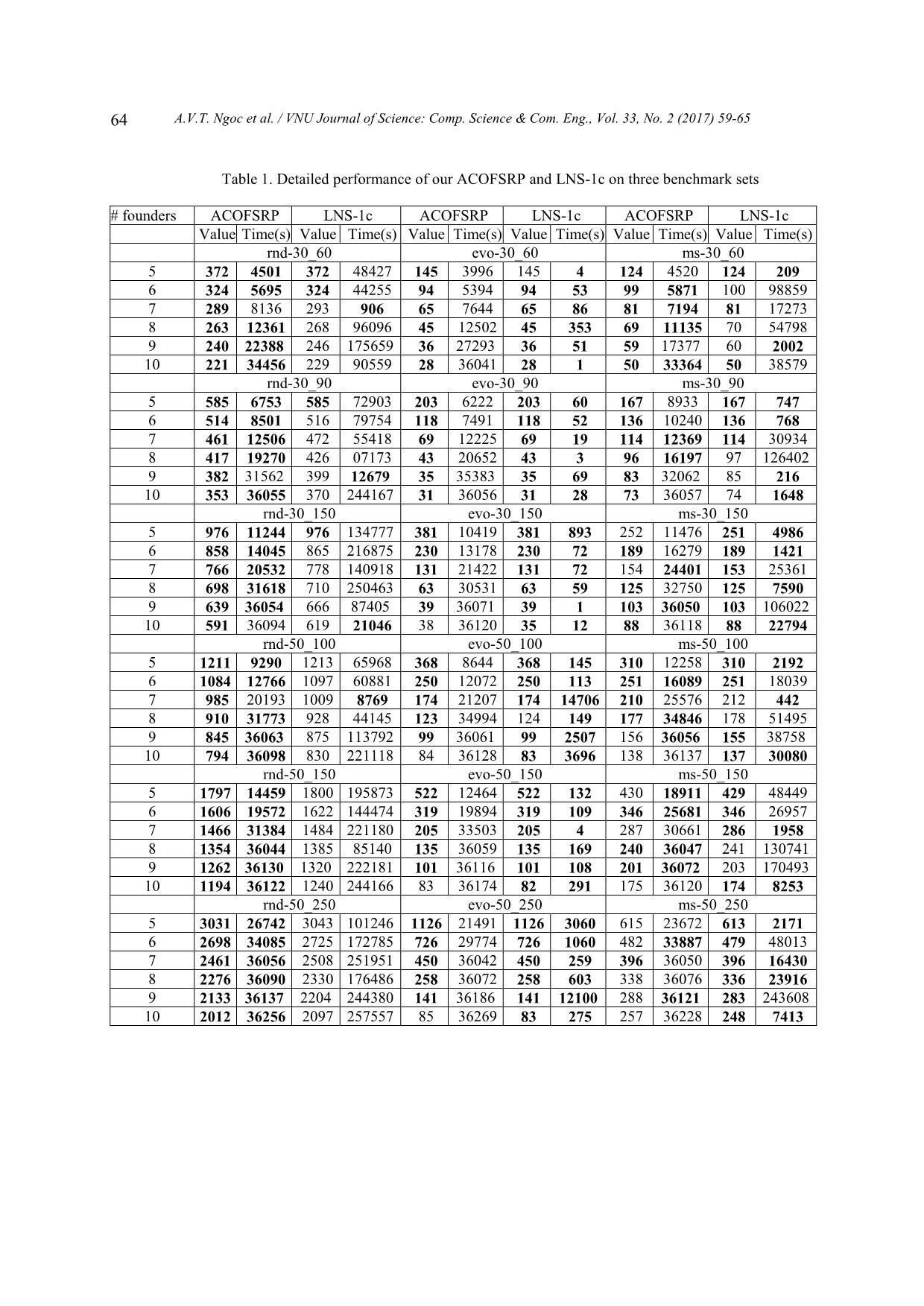
e may ignore 1l V for downstream analysis. 3.2. Large neighborhood search algorithm LNS-1c is empirically considered the best algorithm proposed by far for solving the FSR problem [4]. This algorithm uses the nearest- neighbor search strategy over a large neighborhood of constructed solutions. During searching the neighborhood, the algorithm picks out a set FFfree beforehand, then uses the algorithm Recblock to search for alternative founder sequences in freeFF . Whenever a better solution is found out, LNS- 1c performs local search over neighborhood from scratch. 4 Proposed method 4.1. Ant colony optimization based FSR Ant colony optimization [7] (ACO) is a metaheuristic method simulating how ants in nature find paths from their nest to food sources, which turn out to be a reinforcement learning method. ACO solves optimization problems throughout many episodes, in each of which every ant travels to find solutions based on heuristic information and pheromone matrix containing information learned. The best solution found in the current episode is used to learn (tune ) and go for the next turn. Our proposed method for FSR has input and output as follows: Input: binary matrix C of size mn* representing a recombinant set and k is the number of the founder sequences to be found. Output: binary matrix F of size mk * string representing the founder sequences so that ),( FCBP is minimal. Here, ),( FCBP is the number of breakpoints required to obtain C from F . In general, our ACO based method for FSR works as depicted in Algorithm 1: 4.2. Structure graph for the FSR problem For the sake of visualization, we simulate the FSR problem as the problem of finding paths on a corresponding structure graph (see Figure 2). This structure graph includes a start, an end node and m columns. Each column has k2 vertices, of which each corresponds to a state of the corresponding column in the matrix F of founder sequences. In particularly, each state is a binary string of length k . Each vertex has edges connecting to all ones in the next column. We can see all paths starting from the start to the end node has to go through every column once, at which one state is chosen. Each journey of ants travelling from the start to the end node therefore corresponds to a complete matrix of founder sequences. 4.3. How ants travel on the structure graph When travelling on the structure graph, ants chose a next vertex to visit at random. The A.V.T. Ngoc et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 33, No. 2 (2017) 59-65 62 algorithm is described in pseudo code in Algorithm ??. The probability at which a vertex is chosen is proportional to its level of compatibility to the matrix constructed by ants so far. This level is calculated through heuristic and pheromone information . Particularly, the j vertex in the column i will be visited by an ant with a probability. ][][ ][][ = ,, ,, , lali l jaji jiP Where: • ja, is the heuristic value (see 4.3.1). • ji , is the pheromone information (see 4.3.2). • , are two parameters of an ACO determining the correlation between the heuristic value and the pheromone information. 4.3.1. Heuristic information While constructing the optimal solution, heuristic information is calculated according to the level of compatibility to the matrix that is yielded with the next moves of ants. In more details, when an ant is going to the j vertex in the column i the heuristic information is calculated as follows. ),( 1 =, jFCBP ai ja where: • iC is the matrix of the first i columns of matrix C . • aF is the solution that ant a has built (with 1 i columns). • jFa is the matrix resulted when ant a intends to visit vertex j . To give an example, when 3=i we have the structure graph as in Figure 3. Figure 3. Structure graph when i = 3. 4.3.2. Pheromone information In the FSR problem, we denote ij as the pheromone information of the thj vertex in the column i in the graph. Vertices being visited in the optimal solutions found in every searching phase by ants so far will be learnt such that they are of high priority to be visited in next phases. There are various pheromone updating methods that have been proposed for ACO. We select the Smoothed Max-Min Ant system [8] because it yields the best results in our experiments. In this regard, the pheromone information is updated after each loop as follows: ijijij )(1= where: Tjiif Tjiif max min ij ),( ),( = and T is the optimal solution that ants found after the loop and ),( ji is the vertex j in the column i of the structure graph. 4.4. Improved ACO for FSRP 4.4.1 Ants find solutions synchronously Note that the problem solution space is extremely large, if working independently with Figure 2. Structure graph for the ACO-based founder sequence reconstruction. A.V.T. Ngoc et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 33, No. 2 (2017) 59-65 63 each other ants could hardly to concentrate on potential regions of the searching space. We therefore propose a search strategy for ants as follows: We let ants (in the set Ants) find solutions in parallel. When moving to the next column, instead of letting each ant choose the next vertex to go, we create a new ant set (called NewAnts) to prolong paths created by ants in the set Ants. In particular, if an ant a prolongs the path for an ant a , it means that ant a will go over the similar journey as ant a before moving to the next vertex in the next column. When having NewAnts with the same size as Ants, we move to the next column and repeat such a new ant set building procedure from NewAnts until having a complete solution set. This procedure is depicted in pseudo code in Algorithm 3. For more details, when going from the column 1 i to the column i , each ant NewAntsa will randomly choose an ant Antsa to prolong its path and a vertex j in the column i to move forward. The ant a is chosen with a probability also based on the heuristic and pheromone information, as follows: ][][ ][][ = ,, ,, , lxali lxa jaji jaP 4.4.2. Other improvements Neighborhood search: To lower the probability of missing good solutions while searching, we recommend using the reduced version of the algorithm RecBlock (3.2) to find other better solutions within the vicinity of the best by far solution found by ants. Instead of browsing the whole founder sequences, for each founder in the optimal solution found by far we use RecBlock to find another alternative better one. Searching along two dimensions: With the newly proposed search strategy, ants will quickly converge onto some solution regions, leading to a low diversity of found solutions. To improve this problem, apart from searching forward from the start to the end vertex, we also let ants search backward along the opposite direction (i.e. from the end back to start vertex). The search direction is periodically changed. When searching backward, the complete different heuristic information is used, leading to the potential of finding new solutions. 5. Experimental results We compare our proposed FSR algorithm called ACOFSRP with the best corresponding one by far, i.e. LNS-1c [4] on 3 benchmark data sets, namely rnd (random), evo and ms (each contains 6 test set). All sequences in the first data set is randomly generated while those in the two latter ones are generated according to evolutionary models. All three are used in the study of LNS-1c. We do experiments with the founder sequence length 105,6,7,8,9, k for each of such 3 test sets, leading to a total of 108 tests. We also do experiments with different variants of ACOFSRP by not using either one of two improvements or both on the same three benchmark sets. Experimental results show that ACOFSRP outperforms its two variants, demonstrating the power of two proposed improvements in ACOFSRP (data not shown). Due to the random nature of ACOFSRP, we perform each test 20 times and the run time of each is limited to 10 hours. These numbers are 1 and 72, respectively, in the study of LNS-1c [4]. The program is run on a CPU with 12GB RAM and 4GHz processor. Table ?? shows the detailed performance, in terms of the solution quality (number of required breakpoints) and the running time, of ACOFSRP and LNS-1c on three benchmark data sets. Note that the values for ACOFSRP are the averages of those from 20 running times. A.V.T. Ngoc et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 33, No. 2 (2017) 59-65 64 Table 1. Detailed performance of our ACOFSRP and LNS-1c on three benchmark sets # founders ACOFSRP LNS-1c ACOFSRP LNS-1c ACOFSRP LNS-1c Value Time(s) Value Time(s) Value Time(s) Value Time(s) Value Time(s) Value Time(s) rnd-30_60 evo-30_60 ms-30_60 5 372 4501 372 48427 145 3996 145 4 124 4520 124 209 6 324 5695 324 44255 94 5394 94 53 99 5871 100 98859 7 289 8136 293 906 65 7644 65 86 81 7194 81 17273 8 263 12361 268 96096 45 12502 45 353 69 11135 70 54798 9 240 22388 246 175659 36 27293 36 51 59 17377 60 2002 10 221 34456 229 90559 28 36041 28 1 50 33364 50 38579 rnd-30_90 evo-30_90 ms-30_90 5 585 6753 585 72903 203 6222 203 60 167 8933 167 747 6 514 8501 516 79754 118 7491 118 52 136 10240 136 768 7 461 12506 472 55418 69 12225 69 19 114 12369 114 30934 8 417 19270 426 07173 43 20652 43 3 96 16197 97 126402 9 382 31562 399 12679 35 35383 35 69 83 32062 85 216 10 353 36055 370 244167 31 36056 31 28 73 36057 74 1648 rnd-30_150 evo-30_150 ms-30_150 5 976 11244 976 134777 381 10419 381 893 252 11476 251 4986 6 858 14045 865 216875 230 13178 230 72 189 16279 189 1421 7 766 20532 778 140918 131 21422 131 72 154 24401 153 25361 8 698 31618 710 250463 63 30531 63 59 125 32750 125 7590 9 639 36054 666 87405 39 36071 39 1 103 36050 103 106022 10 591 36094 619 21046 38 36120 35 12 88 36118 88 22794 rnd-50_100 evo-50_100 ms-50_100 5 1211 9290 1213 65968 368 8644 368 145 310 12258 310 2192 6 1084 12766 1097 60881 250 12072 250 113 251 16089 251 18039 7 985 20193 1009 8769 174 21207 174 14706 210 25576 212 442 8 910 31773 928 44145 123 34994 124 149 177 34846 178 51495 9 845 36063 875 113792 99 36061 99 2507 156 36056 155 38758 10 794 36098 830 221118 84 36128 83 3696 138 36137 137 30080 rnd-50_150 evo-50_150 ms-50_150 5 1797 14459 1800 195873 522 12464 522 132 430 18911 429 48449 6 1606 19572 1622 144474 319 19894 319 109 346 25681 346 26957 7 1466 31384 1484 221180 205 33503 205 4 287 30661 286 1958 8 1354 36044 1385 85140 135 36059 135 169 240 36047 241 130741 9 1262 36130 1320 222181 101 36116 101 108 201 36072 203 170493 10 1194 36122 1240 244166 83 36174 82 291 175 36120 174 8253 rnd-50_250 evo-50_250 ms-50_250 5 3031 26742 3043 101246 1126 21491 1126 3060 615 23672 613 2171 6 2698 34085 2725 172785 726 29774 726 1060 482 33887 479 48013 7 2461 36056 2508 251951 450 36042 450 259 396 36050 396 16430 8 2276 36090 2330 176486 258 36072 258 603 338 36076 336 23916 9 2133 36137 2204 244380 141 36186 141 12100 288 36121 283 243608 10 2012 36256 2097 257557 85 36269 83 275 257 36228 248 7413 A.V.T. Ngoc et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 33, No. 2 (2017) 59-65 65 On the random data set ( rnd ), ACOFSRP could procedure solutions better than LNS-1c for 32 among total 36 cases. On-par solutions are observed in the 4 remaining cases. Regarding the running time, ACOFSRP requires shorter time than LNS-1c for 32 cases while longer only for 4 remaining cases. On the data set evo , ACOFSRP is beated by LNS-1c in terms of excution time for all cases. Nevertheless, solutions yielded by ACOFSRP are on-par with those of LNS-1c for 32 out of 36 cases. For the remaining 4 cases, the solution goodness scores by ACOFSRP are worse than those by LNS-1c (The small differences are observed, i.e. up to 3 breakpoints). On the data set ms , ACOFSRP produced solutions are better than and equal to those yielded by LNS-1c for 12 and 10 cases, respectively. Interestingly, among such 22, ACOFSRP requires remarkably shorter runing time than LNS-1c for 12 cases. For the remaining 14 cases, ACOFSRP produce solutions worse than LNS- 1c. ./table_combine_all.tex 6. Conclusion Founder gene sequence reconstruction (FSR) for a given population can be modeled as a combinatorial optimization problem, which has been proven NP-hard. In this paper we propose a novel method based on ant colony optimization algorithms (ACO) coupled with two other important improvements (i.e. local search and back forward search) to solve the founder gene sequence reconstruction problem. Experiments on the benchmark data sets show better or equal results for almost sets when comparing to the best corresponding method, demonstrating the efficacy and future perspectives of our proposed method. Acknowledgments This work has been supported by Vietnam National University, Hanoi (VNU), under Project No. QG.15.21. References [1] G. Tyson, J. Chapman, H. Philip, E. Allen, R. Ram, P. M. Richardson, V. Solovyev, E. M. Rubin, D. Rokhsar, J. F. Banfield, Community structure and metabolism through reconstruction of microbial genomes from the environment, Nature 428 (2004) 37–43. [2] E. Ukkonen, Finding Founder Sequences from a Set of Recombinants, Springer Berlin Heidelberg, Berlin, Heidelberg, 2002, pp. 277–286. [3] A. Roli, C. Blum, Tabu Search for the Founder Sequence Reconstruction Problem: A Preliminary Study, Springer Berlin Heidelberg, Berlin, Heidelberg, 2009, pp. 1035–1042. [4] A. Roli, S. Benedettini, T. Stützle, C. Blum, Large neighbourhood search algorithms for the founder sequence reconstruction problem, Computers Operations Research 39 (2) (2012) pp. 213–224. [5] C. Blum, A. Roli, Metaheuristics in combinatorial optimization: Overview and conceptual comparison, ACM Comput. Surv. 35 (3) (2003) 268–308. [6] P. Rastas, E. Ukkonen, Haplotype inference via hierarchical genotype parsing, in: Proceedings of the 7th International Conference on Algorithms in Bioinformatics, WABI’07, Springer-Verlag, Berlin, Heidelberg, 2007, pp. 85–97. [7] M. Dorigo, T. Stützle, Ant Colony Optimization, Bradford Company, Scituate, MA, USA, 2004. [8] D. Do Duc, H. Hoang Xuan, Smooth and three- levels ant systems: Novel aco algorithms for solving traveling salesman problem, in: Ad. Cont. to the International Conference: IEEE- RIVF 2010, pp. 33–37. G g
File đính kèm:
 ant_colony_optimization_based_founder_sequence_reconstructio.pdf
ant_colony_optimization_based_founder_sequence_reconstructio.pdf

