Varietal evaluation and genetic variability in rice (Oryza sativa L.) genotypes of the Mid-Hill region of Nepal
The objectives of this study were to identify promising rice genotypes
and evaluate the genetic variance and effectiveness of selection of the
rice varieties for several yield attributing traits. A varietal trial of
fifteen rice genotypes was laid out in a randomized complete block
design (RCBD) with three replications in a farmer’s field in
Sundarbazar, Lamjung, Nepal during the rainy season of 2018.
Analysis of variance indicated that all the genotypes showed
significant variation for all the traits considered. The phenotypic
coefficient of variation (PCV) was higher than the genotypic
coefficient of variation (GCV) for all the characteristics being studied
indicating the presence of environmental influence on the traits. High
heritability coupled with high genetic advance as a percent of the
mean was found for days to physiological maturity, number of tillers
per m2, plant height, leaf area, effective tillers per m2, flag leaf area,
test weight, grains per panicle, filled grains per panicle, harvest index,
grain yield, and straw yield indicating that additive gene interaction
is present in their inheritance. Direct selection can be effective for
yield improvement in the populations through selection of these
traits. Cluster analysis based on eighteen traits grouped the fifteen
rice genotypes into four clusters. Cluster I was the largest and
consisted of five genotypes. Radha 11, NR 119, and Sukhadhan-5
were the top performing genotypes having yield potentials of 5.78,
5.49, and 4.89 tons per ha, respectively.

Trang 1

Trang 2

Trang 3
Trang 4
Trang 5

Trang 6
Trang 7
Trang 8

Trang 9
Trang 10
Tải về để xem bản đầy đủ
Tóm tắt nội dung tài liệu: Varietal evaluation and genetic variability in rice (Oryza sativa L.) genotypes of the Mid-Hill region of Nepal

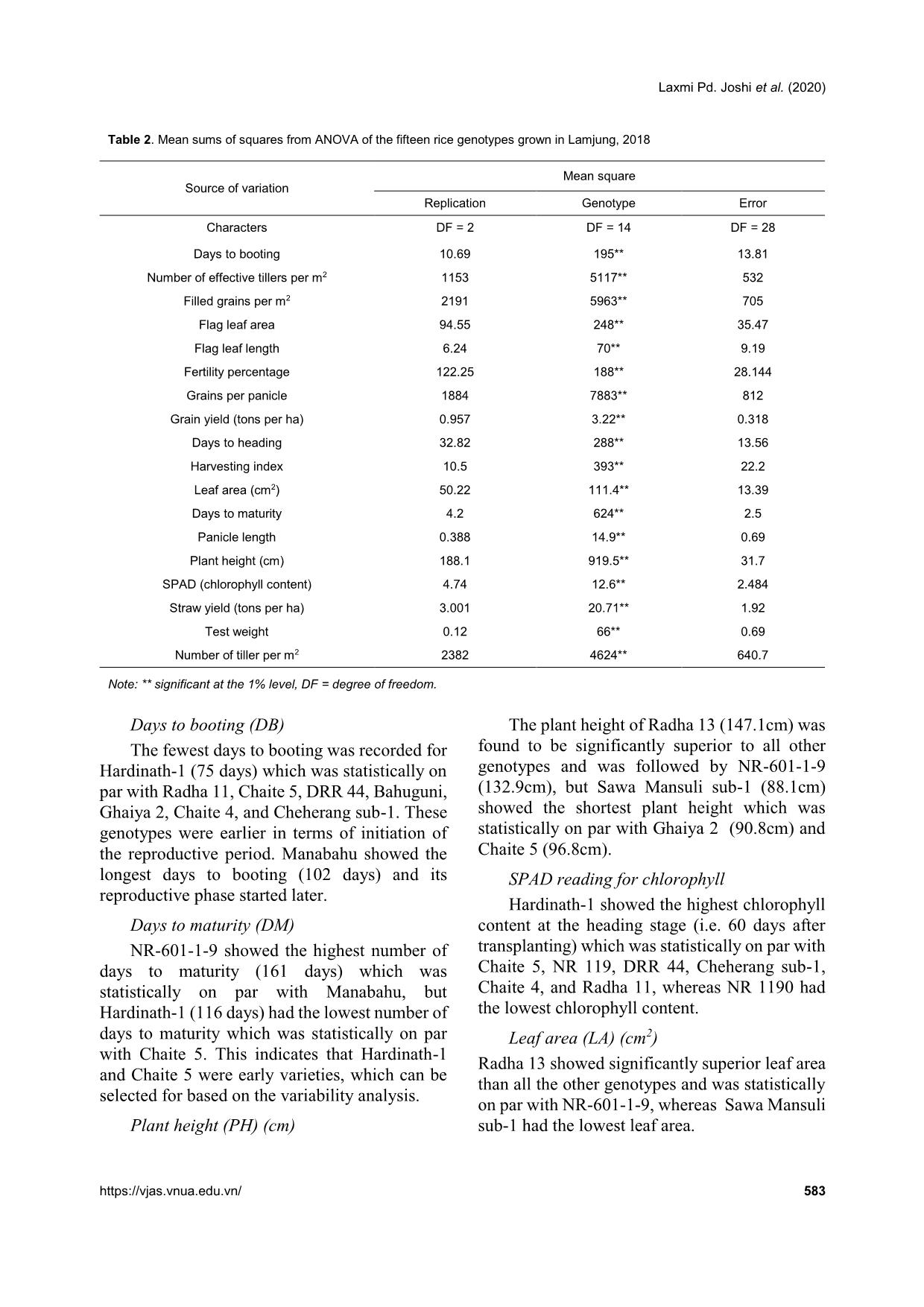
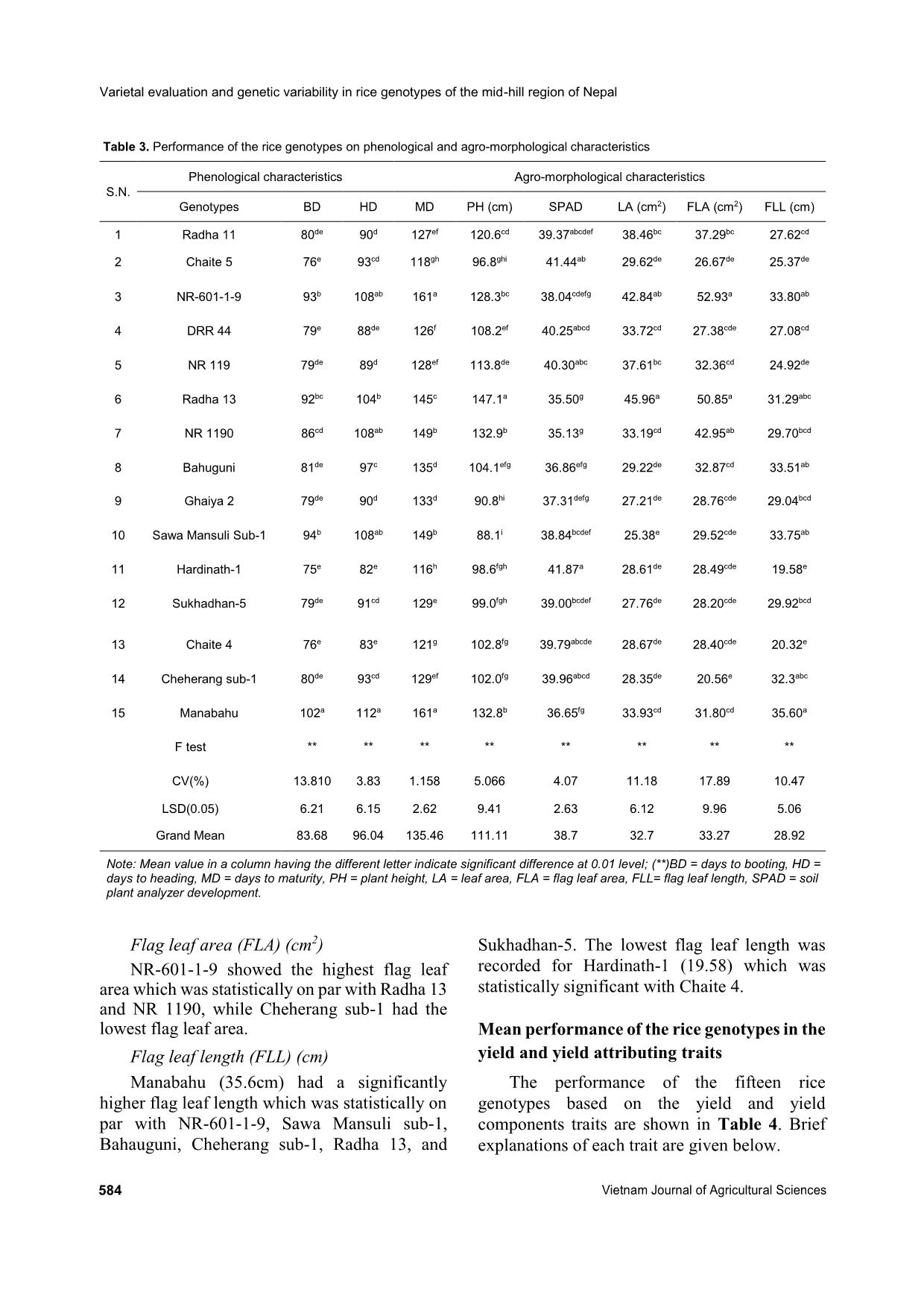
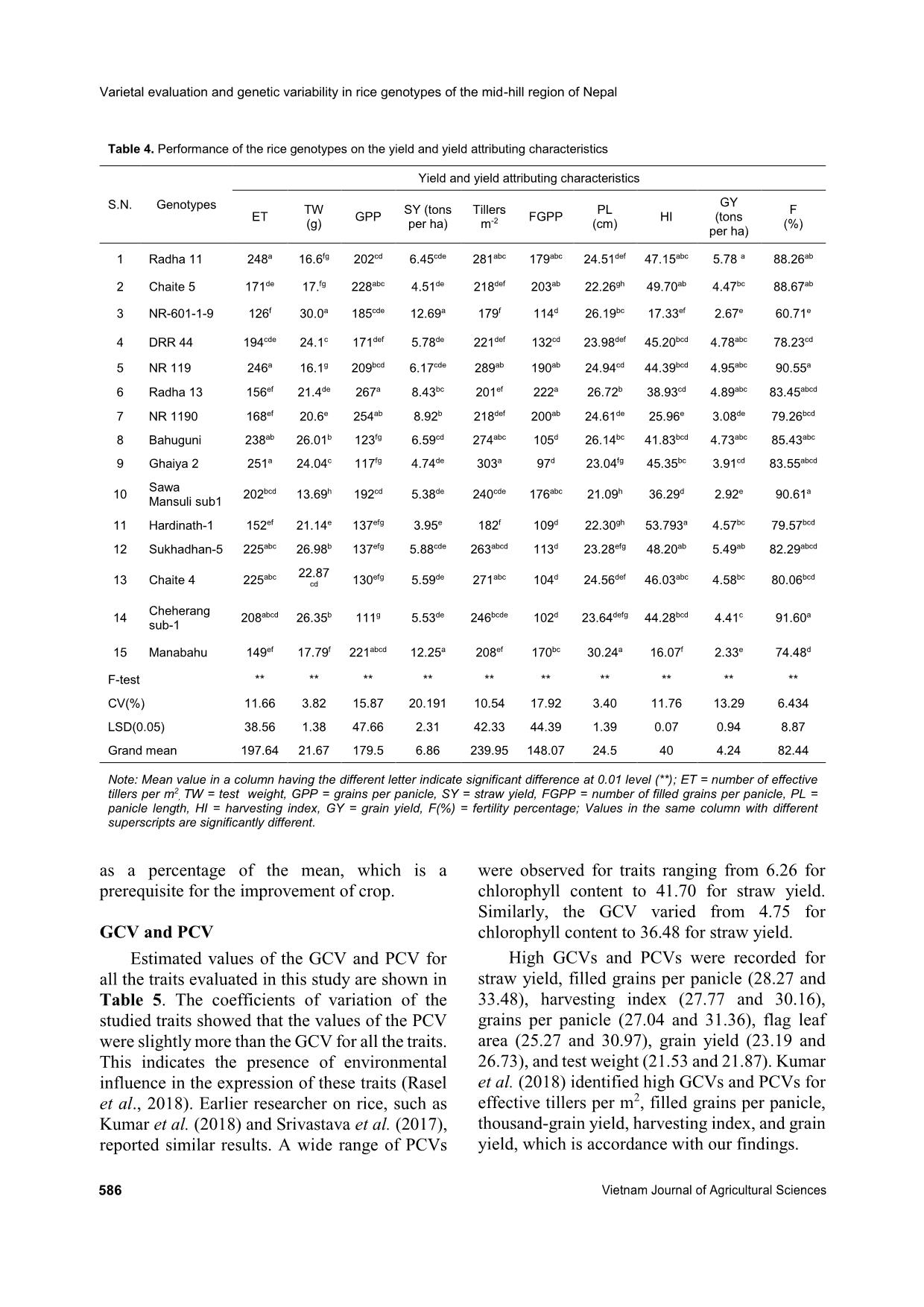
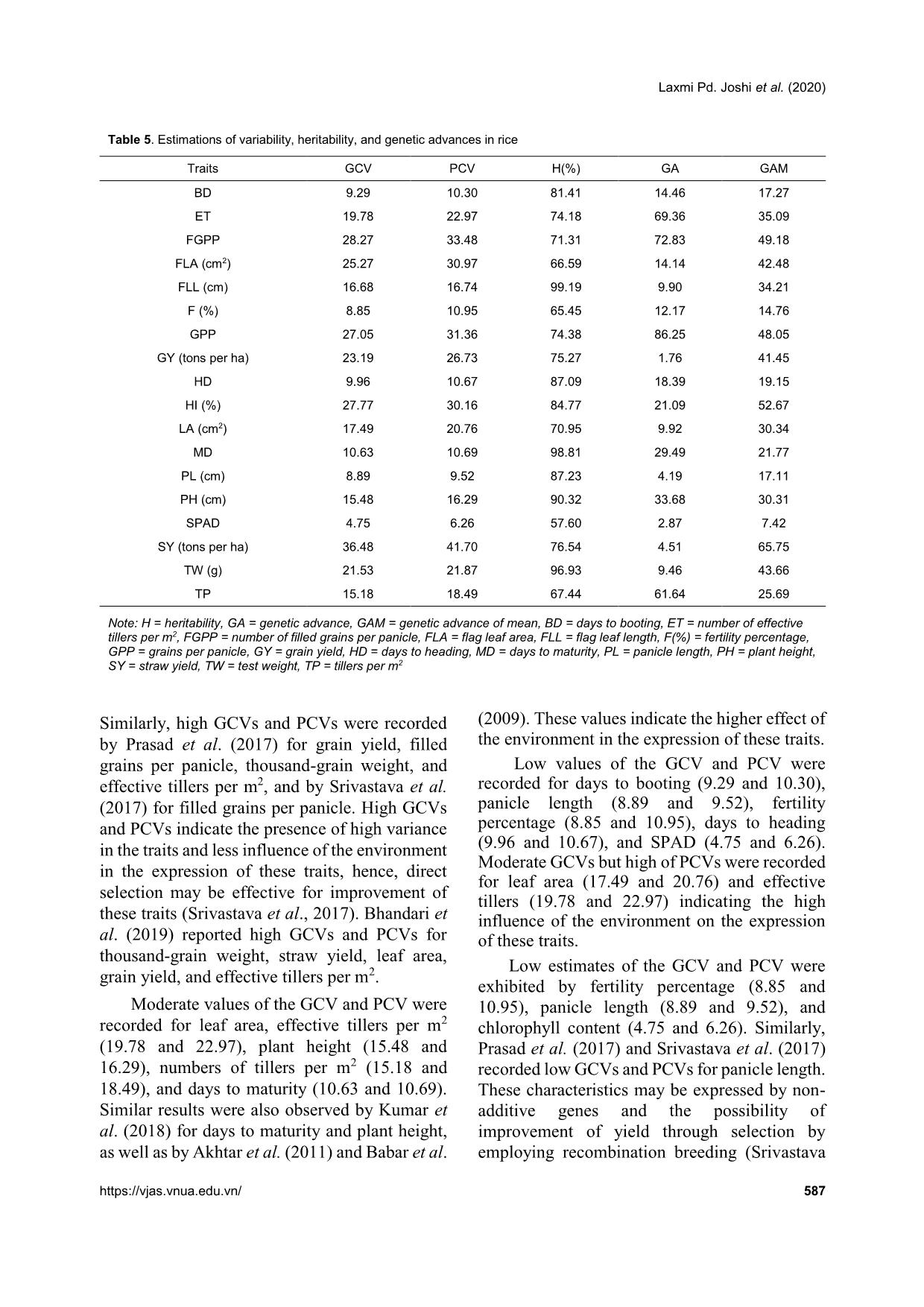
heritability were recorded for days to maturity, days to flowering grain yield, straw yield, and plant height by Roy et al. (2015). High values of broad sense heritability indicate that the traits being studied have the scope to improve through selection. These traits are less influenced by the environment and could be successfully transferred to the next generations if utilized in a hybridization program so that superior genotypic selection can be made through simple selection. Low heritability of traits indicates more influence of the environment in the phenotypic expression of that trait and hence, selection based on progeny or family testing should be done. Genetic advance Genetic advance as a percentage of the mean was found to be highest for straw yield (65.75%) followed by harvesting index (52.67%), filled grains per panicle (49.18%), grains per panicle (48.04%), thousand-grain weight (43.66%), flag leaf area (42.48%), grain yield (41.45%), effective tillers per m2 (35.09%), flag leaf length (42.48%), leaf area (30.34%), plant height (30.31%), tillers per m2 (25.69%), and days to maturity (21.76%). Similar high values of genetic advance as a percentage of the mean were previously recorded by Srivastava et al. (2017) for plant height, flag leaf length, number of grains per panicle, filled grains per panicle, test weight, and grain yield per plant. Medium genetic advance as a percentage of the mean values were found for days to booting (17.23%), panicle length (17.11%), days to heading (19.15%), and fertility percentage (14.75%). Low genetic advance as a percentage of the mean was found for SPAD reading (7.42%). High heritability coupled with high genetic advance as a percentage of the mean were recorded for days to maturity, number of tillers per m2, plant height, leaf area, flag leaf length, effective tillers per m2, grain yield, flag leaf area, test weight, panicle density, grains per panicle, filled grains per panicle, harvesting index, and straw yield. High GCVs and PCVs, heritability, and genetic advance as a percentage of the mean were recorded for straw yield, harvesting index, filled grains per panicle, grains per panicle, thousand- grain weight, flag leaf area, grain yield, effective tillers per m2, and leaf area. These characteristics with high values of the GCV, PCV, and heritability Laxmi Pd. Joshi et al. (2020) https://vjas.vnua.edu.vn/ 589 Table 6. Phenological, agro-morphological, and yield attributing traits of the rice genotypes within and among four clusters Variable Cluster I Cluster II Cluster III Cluster IV Grand centroid BD 79.33 83.25 89.16 84.50 83.68 HD 91.26 94.08 104.50 95.00 96.04 MD 129.60 132.83 143.75 138.83 135.46 PH (cm) 99.77 107.70 127.45 113.54 111.10 SPAD 38.58 39.69 37.18 39.96 38.69 LA (cm2) 28.24 33.79 35.68 35.72 32.70 FLL (cm) 29.02 28.34 30.49 26.69 28.92 FLA (cm2) 27.76 31.64 38.07 40.71 33.27 PL (cm) 24.13 23.63 25.95 24.24 24.50 ET 229.66 223.00 161.50 139.16 197.64 Tillers m2 271.66 258.16 211.66 180.83 239.95 FGPP 104.66 169.42 199.18 111.68 148.07 GPP 124.25 194.10 243.12 161.20 179.50 TW (g) 25.24 17.64 19.28 25.58 21.67 SY (tons ha-1) 5.67 5.94 8.53 8.32 6.86 GY (tons ha-1) 4.62 4.61 3.69 3.62 4.24 HI (%) 45.14 43.25 32.66 35.56 40.03 F (%) 84.58 86.91 81.46 70.14 82.44 Note: BD = days to booting, ET = number of effective tillers per m2, FGPP = number of filled grains per panicle, FLA = flag leaf area, FLL = flag leaf length, F(%) = fertility percentage, GPP = grains per panicle, GY = grain yield, HD = days to heading, MD = days to maturity, PL = panicle length, PH = plant height, SY = straw yield, TW = test weight, TP = tillers per m2 accompanied by high genetic advance as a percentage of the mean might be transmitted to their progenies and therefore, phenotypic selection based on these characteristics would be effective. Panse (1957) stated that high values of heritability with low genetic advance indicated that the heritability was probably due to the effects of non-additive gene action. In general, the characteristics that show high heritability with high genetic advance are controlled by additive gene action (Patil & Lokesha, 2003) and can be improved through simple or progeny selection methods. Selection for the traits having high heritability coupled with high genetic advance is likely to accumulate more additive genes leading to further improvement of their performance (Johnson et al., 1955). The characteristics showing high heritability along with moderate or low genetic advance can be improved by intermating superior genotypes of a segregating population developed from combination breeding. Cluster analysis Cluster analysis of the fifteen rice genotypes in this study showed that the genotypes exhibited considerable genetic variability among themselves by occupying four clusters as shown in Table 6. These rice genotypes were grouped based on phenological, agro-morphological, and yield related traits, namely days to booting, number of effective tillers per m2, filled grains per panicle, flag leaf area, flag leaf length, fertility percentage, grains per panicle, grain yield, days to heading, harvesting index, leaf area, days to maturity, panicle length, plant height, SPAD reading, straw yield, test weight, and number of tillers per m2. Cluster analysis showed that cluster I was comprised of five genotypes, cluster II consisted of four genotypes, cluster III was comprised of four genotypes, and cluster IV consisted of two genotypes (Figure 1). Cluster II genotypes were characterized as having the highest fertility, Varietal evaluation and genetic variability in rice genotypes of the mid-hill region of Nepal 590 Vietnam Journal of Agricultural Sciences smallest palnicle lengths, and lowest thousand- grain weights. Cluster III genotypes had the lowest SPAD readings at the time of flowering, and the longest number of days for booting, anthesis, and harvesting. It was also characterized by having the longest panicle lengths, largest numbers of grains per panicle, largest numbers of filled grains per panicle, and highest straw yields but lowest harvesting indicies. Cluster IV contained the genotypes with the highest SPAD readings at the time of flowering, largest leaf areas, flag leaf areas, and thousand-grain weights, in addition to the lowest flag leaf lengths, and lowest numbers of total and effective tillers per m2. Cluster I had the genotypes that were earliest in terms of booting, heading, and days to maturity, lowest leaf areas, lowest flag leaf areas, and lowest numbers of filled grains per panicle and grains per panicle. It was comprised of genotypes with the highest grain yields, harvesting indicies, and numbers of effective tillers per panicle. Conclusions The analysis of variance study showed the presence of adequate phenotypic variability among the tested genotypes for all the traits in this study. From the mean performance analysis, it was found that Radha 11 can be selected for higher grain yield. Radha 13 can be selected for grains per panicle, tallest plant height, and leaf area. Ghaiya 2 can be selected for the largest number of panicles per m2 and effective tillers per m2. Hardinath-1 can be selected for maximum flag leaf area, chlorophyll content, and earlier heading and maturity, while the local landrace (Manabahu) also had a high values for panicle length, straw yield, flag leaf length, and maximum days to heading, booting, and maturity, and can be selected for these traits. The PCV was greater than the GCV for all the traits being studied, meaning that some degree of environmental influence was present in the expression of these traits. A small difference between the CGV and PCV suggests less influence of the environment on the expression of the traits. High GCVs and PCVs, higher broad sense heritability, and higher GAM of the traits Figure 1. Cluster analysis showing the presence of variability among the tested genotypes and groupings of four distinct genotype clusters based on the phenological, agromorphological, and yield attributing characteristics Observations S im ila ri ty N R -6 0 1 -1 -9 H a rd in a th -1 R a d h a 1 3 N R 1 1 9 0 M a n a b a h u C h a it e 5 R a d h a 1 1 N R 1 1 9 S a w a M a n su li su b -1 D R R 4 4 S u k h a d h a n 5 G h a iy a 2 S u k h a d h a n -5 C h e h e ra n g s u b -1 C h a it e 4 B a h u g u n i 37.57 58.38 79.19 100.00 Laxmi Pd. Joshi et al. (2020) https://vjas.vnua.edu.vn/ 591 straw yield, filled grains per panicle, harvesting index, grains per panicle, flag leaf area, grain yield, thousand-grain weight, effective tillers per m2, and leaf area indicates that there is less influence of the environment in the expression of these traits. A higher proportion of variability is heritable and improvement can be made for these traits in the next generation through direct selection. High broad sense heritability and genetic advance for these traits indicate the presence of additive gene interaction in the expression of quantitative traits and showed more opportunity for selection of these traits for improvements in yield. Cluster analysis showed the presence of considerable genetic variability among the genotypes for further improvement of the genotypes. References Abebe T. S., Alamerew S. & Tulu L. (2017). Genetic variability, heritability and genetic advance for yield and its related traits in rainfed lowland rice (Oryza sativa L.) genotypes at Fogera and Pawe, Ethiopia. Advanced Crop Science Technology. 5: 272. Adhikari B. N., Joshi B. P., Shrestha J. & Bhatta N. R. (2018). Genetic variability, heritability, genetic advance and correlation among yield and yield components of rice (Oryza sativa L.). Journal of Agriculture and Natural Resources. 1(1): 149-160. Akhter N., Nazir M. F., Rabnawz A., Mahmood T., Safdar M. E., Asif M. & Rehman A. (2011). Estimation of heritability, correlation and path coefficient analysis in fine grain rice (Oryza sativa). The Journal of Animal and Plant Sciences. 21(4): 660-664. Akinwale M. G., Gregorio G., Nwilene F. & Akinyele B. O. (2011). Heritability and correlation coefficient analysis for yield and its components in rice (Oryza sativa L.). African Journal of Plant Science. 5(3): 207- 212. Babar M., Khan A. & Arif A. (2009). Path analysis of some leaf and panicle traits affecting grain yield in double haploid lines of rice (Oryza sativa L.). World Journal of Agricultural Sciences. 45(4): 245-252. Bhandari K., Poudel A., Sharma S., Kandel B. P. & Upadhyay K. (2019). Genetic variability, correlation and path analysis of rice genotypes in rainfed condition at Lamjung, Nepal. RJOAS. 8(92): 274-280. DOI: 10.18551/rjoas.2019-08.30. Chaudhary L. B. & Prasad B. (1968). Genetic variation and heritability of quantitative characters in Indian mustard (Brassica juncea). Indian Journal of Agricultural Science. 38: 820-825. Deshmukh S. N., Basu M. S. & Reddi P. S. (1986). Genetic variability, character association and path coefficient of quantitative traits in Virginia bunch varieties of groundnut. Indian Journal of Agriculture. 56: 816-821. Falconer D. S. & Mackay T. F. C. (1996). An introduction to Quantitative Genetics (4th ed.). London, L; Prentice Hall. Gyawali S., Poudel A. & Poudel S. (2018). Genetic variability and association analysis in different rice genotypes in Mid-Hill of western Nepal. Acta Scientific Agriculture. 2(9): 69-76. Johnson H. W., Robinson H. F. & Comstock R. E. (1955). Estimates of genetic and environmental variability in soybean. Agronomy Journal. 47: 314-318. Kandel B. P., Adhikari N. R., Poudel A. & Tripathi M. P. (2019). Genetic variability estimates of hybrid maize genotypes in inner terai of Nepal. Azarian Journal of Agriculture. 6(6): 164-170. Kumar S., Chauhan M. P. & Tomar A. (2018). Coefficient of variation (GCV & PCV), heritability and genetic advance analysis for yield contributing characters in rice (Oryza sativa L.). Journal of Pharmacognosy and Phytochemistry. 7(3): 2161-2164. MoAD (2016). Statistical Information on Nepalese Agriculture, Ministry of Agriculture Development. NARC (2009). Annual report of Nepal Agriculture Research Council (2007/2008). Retrieved on October 5, 2019 at r_see&id=6638. Pandey P., Anurag J. P. & Tiwari D. K. (2009). Genetic variability, diversity and association of quantitative traits with grain yield in rice (Oryza sativa L.). Journal of Bio-Science. 17(1): 77-82. Panse V. G. (1957). Genetics of quantitative characters in relation to plant breeding. Indian Journal of Genetics. 17: 318-328. Panwar A., Dhaka R. P. S. & Kumar V. (2007). Genetic variability and heritability studies in rice. Advances in Plant Sciences. 20(1): 47-49. Patil M. K. & Lokesha R. (2003). Estimation of Genetic variability, heritability, genetic advance, correlations and path analysis in advanced mutant breeding lines of Sesame (Sesamum indicum L.). Journal of Pharmacognosy and Natural Products. 4151. Prasad R. K., Radhakrishna K. V. & Bhave M. H. V. (2017). Genetic variability, heritability and genetic advance in boro rice (Oryza sativa L.) germplasm. International Journal of Current Microbiology and Applied Sciences. 6(4): 1261-1266. Rasel M. L., Hassan M., Hoqu I. U. & Saha S. R. (2018). Estimation of genetic variability, correlation and path coefficient analysis in local landraces of rice (Oryza sativa L.) for the improvement of salinity tolerance. Journal of the Bangladesh Agricultural University. 16(1): 41-46. Varietal evaluation and genetic variability in rice genotypes of the mid-hill region of Nepal 592 Vietnam Journal of Agricultural Sciences Robinson H. F., Cornstock R. E. & Harvey P. H. (1949). Estimates of heritability and degree of dominance in corn. Agronomy Journal. 41: 353-359. Srivastava N., Babu G. S. & Singh O. N. (2017). Genetic variation, heritability and diversity analysis of exotic upland rice (Oryza sativa L.) germplasms based on quantitative traits. The Pharma Innovation Journal. 6(12): 316-320. Yadav R. K., Gurung R., Dhakal R., Adhikari A. R., Gautam S., Ghimire K. H. Sthapit B. R. (2019). On- farm diversity assessment and participatory varietal evaluation of cold-tolerant rice in Mid-hills of Nepal. Journal of Crop Science and Biotechnology. 22: 403- 414. DOI: 10.1007/s12892-018-0167-0. .
File đính kèm:
 varietal_evaluation_and_genetic_variability_in_rice_oryza_sa.pdf
varietal_evaluation_and_genetic_variability_in_rice_oryza_sa.pdf

