Liver segmentation on a variety of computed tomography (CT) images based on convolutional neural networks combined with connected components
Liver segmentation is relevant for several clinical applications. Automatic liver segmentation
using convolutional neural networks (CNNs) has been recently investigated. In this paper, we propose a
new approach of combining a largest connected component (LCC) algorithm, as a post-processing step,
with CNN approaches to improve liver segmentation accuracy. Specifically, in this study, the algorithm
is combined with three well-known CNNs for liver segmentation: FCN-CRF, DRIU and V-net. We
perform the experiment on a variety of liver CT images, ranging from non-contrast enhanced CT images
to low-dose contrast enhanced CT images. The methods are evaluated using Dice score, Haudorff
distance, mean surface distance, and false positive rate between the liver segmentation and the ground
truth. The quantitative results demonstrate that the LCC algorithm statistically significantly improves
results of the liver segmentation on non-contrast enhanced and low-dose images for all three CNNs. The
combination with V-net shows the best performance in Dice score (higher than 90%), while the DRIU
network achieves the smallest computation time (2 to 6 seconds) for a single segmentation on average.
The source code of this study is publicly available at
Trang 1
Trang 2
Trang 3

Trang 4

Trang 5

Trang 6

Trang 7

Trang 8
Trang 9

Trang 10
Tải về để xem bản đầy đủ
Tóm tắt nội dung tài liệu: Liver segmentation on a variety of computed tomography (CT) images based on convolutional neural networks combined with connected components
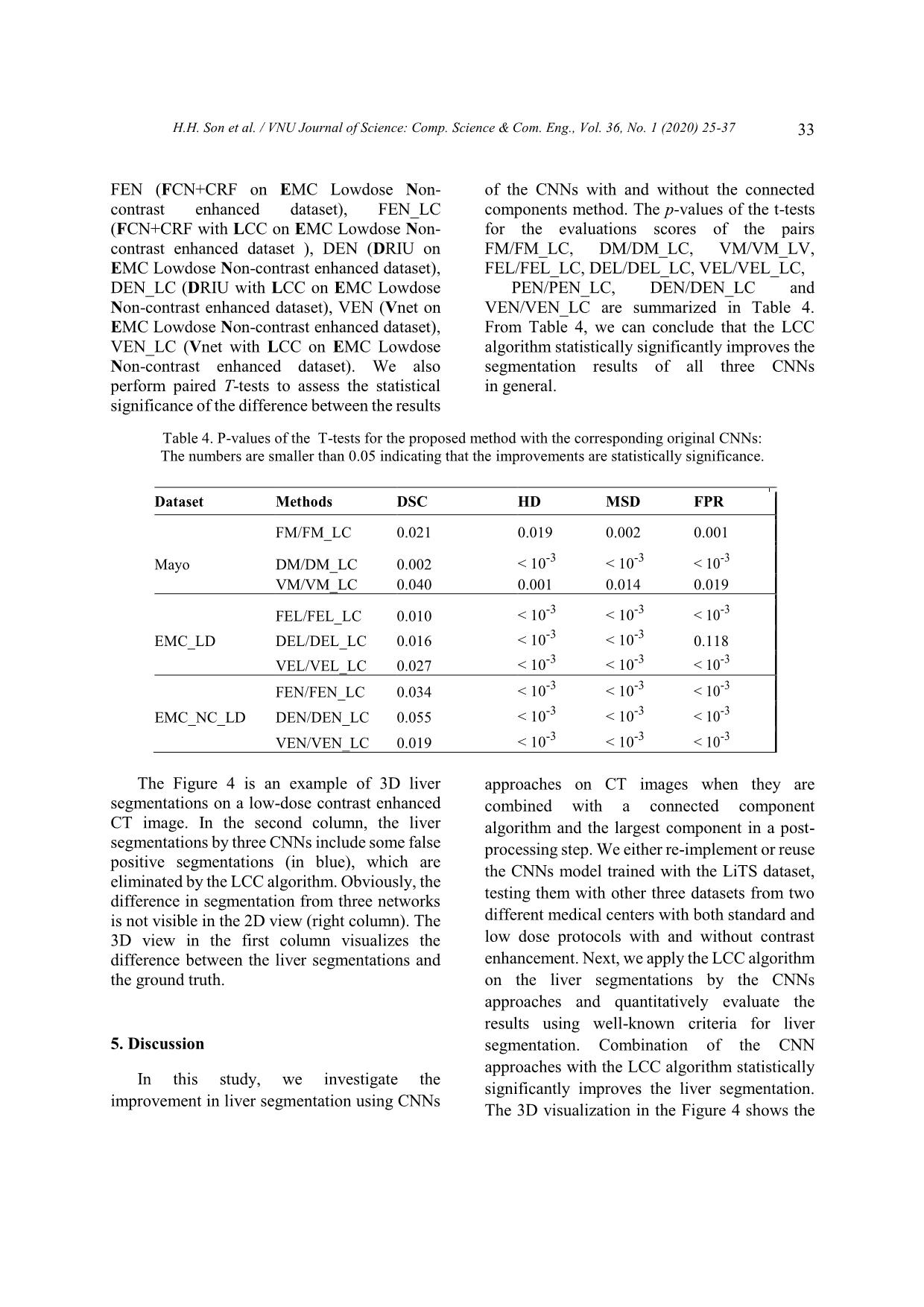
een the results of the CNNs with and without the connected components method. The p-values of the t-tests for the evaluations scores of the pairs FM/FM_LC, DM/DM_LC, VM/VM_LV, FEL/FEL_LC, DEL/DEL_LC, VEL/VEL_LC, PEN/PEN_LC, DEN/DEN_LC and VEN/VEN_LC are summarized in Table 4. From Table 4, we can conclude that the LCC algorithm statistically significantly improves the segmentation results of all three CNNs in general. Table 4. P-values of the T-tests for the proposed method with the corresponding original CNNs: The numbers are smaller than 0.05 indicating that the improvements are statistically significance. Dataset Methods DSC HD MSD FPR FM/FM_LC 0.021 0.019 0.002 0.001 Mayo DM/DM_LC 0.002 < 10 -3 < 10 -3 < 10 -3 VM/VM_LC 0.040 0.001 0.014 0.019 FEL/FEL_LC 0.010 < 10 -3 < 10 -3 < 10 -3 EMC_LD DEL/DEL_LC 0.016 < 10 -3 < 10 -3 0.118 VEL/VEL_LC 0.027 < 10 -3 < 10 -3 < 10 -3 FEN/FEN_LC 0.034 < 10-3 < 10-3 < 10-3 EMC_NC_LD DEN/DEN_LC 0.055 < 10 -3 < 10 -3 < 10 -3 VEN/VEN_LC 0.019 < 10-3 < 10-3 < 10-3 p The Figure 4 is an example of 3D liver segmentations on a low-dose contrast enhanced CT image. In the second column, the liver segmentations by three CNNs include some false positive segmentations (in blue), which are eliminated by the LCC algorithm. Obviously, the difference in segmentation from three networks is not visible in the 2D view (right column). The 3D view in the first column visualizes the difference between the liver segmentations and the ground truth. 5. Discussion In this study, we investigate the improvement in liver segmentation using CNNs approaches on CT images when they are combined with a connected component algorithm and the largest component in a post- processing step. We either re-implement or reuse the CNNs model trained with the LiTS dataset, testing them with other three datasets from two different medical centers with both standard and low dose protocols with and without contrast enhancement. Next, we apply the LCC algorithm on the liver segmentations by the CNNs approaches and quantitatively evaluate the results using well-known criteria for liver segmentation. Combination of the CNN approaches with the LCC algorithm statistically significantly improves the liver segmentation. The 3D visualization in the Figure 4 shows the H.H. Son et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 36, No. 1 (2020) 25-37 34 improvements in a segmentation example. We also conclude that the FCN combined with conditional random forest method does not fully eliminate the isolated false positive segmentation. This can be explained by the fact that the CRF only examines inter-slice correlation of the segmentations, while the liver segmentation should be connected in 3D as one organ. From Figure 3, we can also conclude that the CNNs work better with the regular dose contrast enhanced CT images while most improvements by the LCC occur with the low- dose CT image. This may improve when more low dose images are included in the training stage. We refrained from adding more data in the training stage. In our opinion, while retraining CNNs network is a very “expensive” way of research, reusing the shared works and improving the result using “inexpensive” techniques is a reasonable approach to promote research results to practical application. We also can see from Table 3 and Figure 3 that V-net combined with the LCC generally perform better than other methods. This confirms findings from Milletari et al. (2016) [13], which show that 3D segmentation approaches use inter-slice information and thus may improve segmentation accuracy. However, Table 3 also demonstrates that the 3D nature of the V-net leads to more computation time and requires more memory. These factors may limit its potential to be used in clinical practices that require very fast processing such as intra operation of liver RFA. Note that in our experiment, we already manually cropped the liver volume to avoid the redundancy while current CT scans in clinical practice may have hundreds of slices. A fast, automatic liver detection method may be beneficial for those cases to extract the region of interest while reducing the processing time. Although the LCC shows to be effective for liver segmentation, it still presents challenges. The LCC can only remove false positive segmentations, which are isolated from the main liver segmentation, and thus cannot get rid of false positive segmentations connected with the main part, or fill in missing parts. More advanced segmentation methods, such as level set and graph-cuts, may further improve the smoothing on the surface of the liver, since they can embed and model liver shape and curvature information. Thus, the precise liver surface segmentation needs to be further investigated. Perhaps, subsequent studies may use data sharing to utilize more data in the training stage. While data sharing is currently challenging due to administrative procedures and privacy concerns, data-augmentation research directions could help enrich the training data pools. There are some limitations in our study. First, we only use 10 contrast enhanced CT, 15 low-dose contrast enhanced CT, and 15 low- dose non-contrast enhanced CT from two medical centers for evaluating the methods. Nevertheless, we assume that the images from other medical centers will yield similar results as those in this study. Second, the training dataset for the CNNs does not include low-dose CT images, resulting in poor performance with the EMC dataset. However, while investigating to improve the CNNs with more dataset in the training stage is not the main purpose of our research, we believe that adding low-dose CT images may improve the segmentation results. The improvement may be limited due to effects of the low-dose noise on the image quality. A noise removal CNN network combined with the current CNNs may be a more effective approach to improve the liver segmentation. Third, there have been several other variants of CNNs for liver segmentation that have achieved adequate results [17,23-27]. However, as pixel classification based methods, these CNNs may contain mis-classification parts and may likely benefit as well from post-processing methods such as the LCC. F H.H. Son et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 36, No. 1 (2020) 25-37 35 Figure 4. Example of 3D liver segmentations by the three CNNs on a low-dose contrast enhanced CT image. The first raw is segmentations by FCN, the second one is by DRIU and the last one is by V-net. The first column contains the liver segmentations using with LCC (green) and the ground truth (red), the second column illustrates the raw liver segmentation from the CNNs (blue) overlapped by the segmentation after post processing, and the last column is the final 2D liver segmentations on 2D CT slice of the liver. 6. Conclusion In this paper, we present our work on improving liver segmentation for CNN based approaches using LCC algorithm. Experiments are performed with three well-known CNN architectures and with retrained or reused trained models. We evaluate three datasets from two different medical centers with regular contrast enhanced CT image and both contrast and non- contrast enhancement of low-dose image. The quantitative evaluation results show that LCC statistically significantly improves the liver segmentation accuracy of the CNNs, while maintaining the processing time of less than 10 seconds in total for all of the networks, including the LCC processing time of less than a second. In our study, we find that V-net combined with the LCC achieves a Dice score of approximately 94%, which is comparable to other state of the H.H. Son et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 36, No. 1 (2020) 25-37 36 art methods. We believe that with the current development of CNN-based approach research, the liver segmentation using CNNs has a high potential to be applied in the clinical practice soon. Acknowledgments This work has been supported by VNU University of Engineering and Technology under project number CN 18.03. We would like to thank Mayo Clinical for supporting us their data. We also would like to thank NVIDIA for their aid of a graphics hardware unit. Reference [1] K.A. McGlynn, J.L. Petrick, W.T. London, Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clinics in liver disease 19(2) (2015) 223-238. [2] M. Mohammadian, N. Mahdavifar, A. Mohammadian-Hafshejani, H. Salehiniya, Liver cancer in the world: epidemiology, incidence, mortality and risk factors, World Cancer Res J. 5(2) (2018) e1082. [3] T.T. Hong, N. Phuong Hoa, S.M. Walker, P.S. Hill, C. Rao, Completeness and reliability of mortality data in Viet Nam: Implications for the national routine health management information system, PloS one 13(1) 2018) e0190755. https://doi.org/10.1371/journal.pone.0190755. [4] T. Pham, L. Bui, G. Kim, D. Hoang, T. Tran, M. Hoang, Cancers in Vietnam-Burden and Control Efforts: A Narrative Scoping Review. Cancer Control 26(1) (2019) 1073274819863802. [5] M. Borner, M. Castiglione, J. Triller, H.U. Baer, M. Soucek, L. Blumgart, K. Brunner, Arena: Considerable side effects of chemoembolization for colorectal carcinoma metastatic to the liver, Annals of oncology 3(2) (1992) 113-115. [6] K. Memon, R.J. Lewandowski, L. Kulik, A. Riaz, M.F. Mulcahy, R. Salem, Radioembolization for primary and metastatic liver cancer, In Seminars in radiation oncology, WB Saunders. 21(4) (2011) 294-302. [7] I. Gory, M. Fink, S. Bell, P. Gow, A. Nicoll, V. Knight, W. Kemp, Radiofrequency ablation versus resection for the treatment of early stage hepatocellular carcinoma: A multicenter Australian study, Scandinavian journal of gastroenterology 50(5) (2015) 567-576. [8] H.M. Luu, C. Klink, W. Niessen, A. Moelker, T. Van Walsum, Non-rigid registration of liver CT images for CT-guided ablation of liver tumors. PloS one, 11(9) 92016) e0161600. [9] G. Gunay, M.H. Luu, A. Moelker, T. Van Walsum, S. Klein, Semiautomated registration of pre‐and intraoperative CT for image‐guided percutaneous liver tumor ablation interventions, Medical physics 44(7) (2017) 3718-3725. [10] A. Gotra, L. Sivakumaran, G. Chartrand, N. Vu, F. Vandenbroucke-Menu, C. Kauffmann, A. Tang, Liver segmentation: indications, techniques and future directions, Insights into imaging 8(4) (2017) 377-392. https://doi.org/10.1007/s13244-017- 0558-1. [11] T. Heimann, B. Van Ginneken, M.A. Styner, Y. Arzhaeva, V. Aurich, C. Bauer, F. Bello, Comparison and evaluation of methods for liver segmentation from CT datasets, IEEE transactions on medical imaging 28(8) (2009) 1251-1265. [12] O. Ronneberger, P. Fischer, T. Brox, U-net: Convolutional networks for biomedical image segmentation, In International Conference on Medical image computing and computer-assisted intervention, Springer, Cham, 2015, pp. 234-241. [13] F. Milletari, N. Navab, S.A. Ahmadi, October, V-net: Fully convolutional neural networks for volumetric medical image segmentation, In 2016 Fourth International Conference on 3D Vision (3DV) IEEE, 2016, pp. 565-571. [14] P.F. Christ, F. Ettlinger, F. Grün, M.A. Elshaera, J. Lipkova, S. Schlecht, M. Rempfler, Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks, arXiv preprint arXiv:1702.05970, 2017. [15] P.F. Christ, M.E.A. Elshaer, F. Ettlinger, S. Tatavarty, M. Bickel, P. Bilic, H. Sommer, Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham, 2016, pp. 415-423. [16] H. Meine, G. Chlebus, M. Ghafoorian, I. Endo, A. Schenk, Comparison of U-net-based Convolutional Neural Networks for Liver Segmentation in CT. arXiv preprint arXiv, 2018, pp. 1810.04017. [17] X. Li, H. Chen, X. Qi, Q. Dou, C.W. Fu, P.A. Heng, H-DenseUNet: hybrid densely connected H.H. Son et al. / VNU Journal of Science: Comp. Science & Com. Eng., Vol. 36, No. 1 (2020) 25-37 37 UNet for liver and tumor segmentation from CT volumes, IEEE transactions on medical imaging, 37(12) (2018) 2663-2674. [18] M. Bellver, K.K. Maninis, J. Pont-Tuset, X. Giró- i-Nieto, J. Torres, L. Van Gool, Detection-aided liver lesion segmentation using deep learning, ArXiv preprint arXiv:1711.11069, 2017. [19] H.S. Hoang, C.P. Pham, D. Franklin, T. Van Walsum, M.H. Luu, An Evaluation of CNN-based Liver Segmentation Methods using Multi-types of CT Abdominal Images from Multiple Medical Centers, In 2019 19th International Symposium on Communications and Information Technologies (ISCIT), IEEE, September, 2019, pp. 20-25. [20] H. Samet, M. Tamminen, Efficient component labeling of images of arbitrary dimension represented by linear bintrees, IEEE Transactions on Pattern Analysis and Machine Intelligence, 10(4) (1988) 579-586. [21] P. Bilic, P.F. Christ, E. Vorontsov, G. Chlebus, H. Chen, Q. Dou, S. Kadoury, The liver tumor segmentation benchmark (lits), ArXiv preprint arXiv, 2019, 1901.04056. [22] H.M. Luu, A. Moelker, S. Klein, W. Niessen, T. Van Walsum, Quantification of nonrigid liver deformation in radiofrequency ablation interventions using image registration, Physics in Medicine & Biology 63(17) (2018) 175005. [23] A.A. Novikov, D. Major, M. Wimmer, D. Lenis, K. Bühler, Deep Sequential Segmentation of Organs in Volumetric Medical Scans, IEEE transactions on medical imaging, 2018. [24] Y. Huo, J.G. Terry, J. Wang, S. Nair, A. Lasko, B.I. Freedman, B.A. Landman, Fully Automatic Liver Attenuation Estimation combing CNN Segmentation and Morphological Operations, Medical physics, 2019. [25] N. Gruber, S. Antholzer, W. Jaschke, C. Kremser, M. Haltmeier, A Joint Deep Learning Approach for Automated Liver and Tumor Segmentation, ArXiv preprint arXiv, 2019, pp. 1902.07971. [26] S. Chen, K. Ma, Y. Zheng, Med3D: Transfer Learning for 3D Medical Image Analysis, ArXiv preprint arXiv, 2019, pp. 1904.00625. [27] W. Tang, D. Zou, S. Yang, J. Shi, DSL: Automatic Liver Segmentation with Faster R-CNN and DeepLab, In International Conference on Artificial Neural Networks, Springer, Cham, 2018, pp. 137-147. O -
File đính kèm:
 liver_segmentation_on_a_variety_of_computed_tomography_ct_im.pdf
liver_segmentation_on_a_variety_of_computed_tomography_ct_im.pdf

