Expression of Zinc Finger Protein Zat12 from Arabidopsis thaliana in Escherichia coli
The C2H2 zinc finger protein ZAT12 has been classified as a plant
core abiotic stress response gene in the early response to multiple
stresses. ZAT12 links the iron deficiency and oxidative stress
responses through the direct interaction with/and negative regulation
of a central regulator - FIT. For further research on the regulation of
the ZAT12 protein in planta, a huge quantity of ZAT12 proteins is
required to inject into mice for the generation of ZAT12 antiserum.
In this study, the gene encoding the ZAT12 protein from Arabidopsis
thaliana was cloned into the expression vector - pETBlue-2 and then
overexpressed in E. coli T7. A high expression level was indicated
by SDS-PAGE. Immunoblot demonstrated successful expression
using a bacterial expression system.

Trang 1

Trang 2
Trang 3

Trang 4
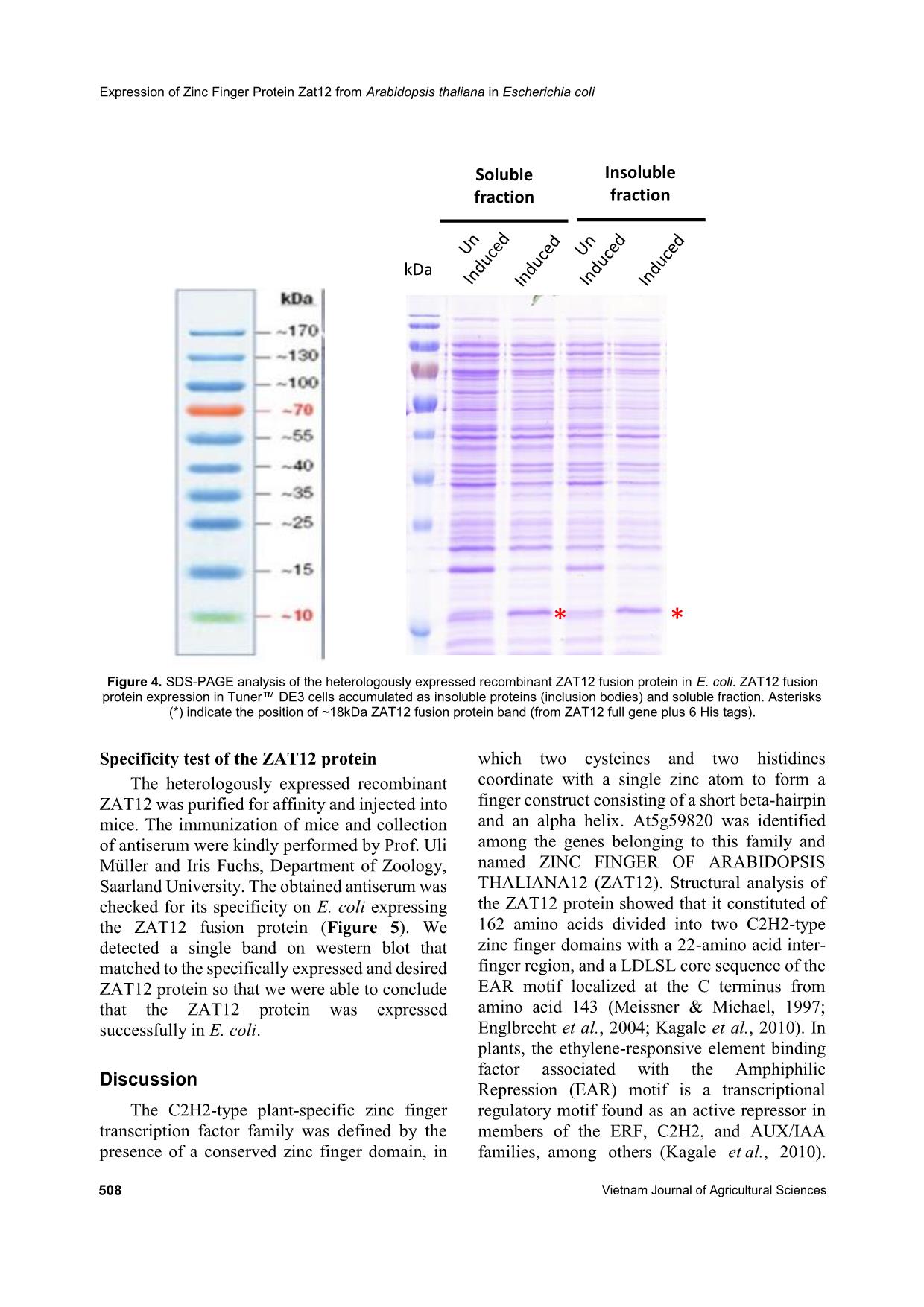
Trang 5
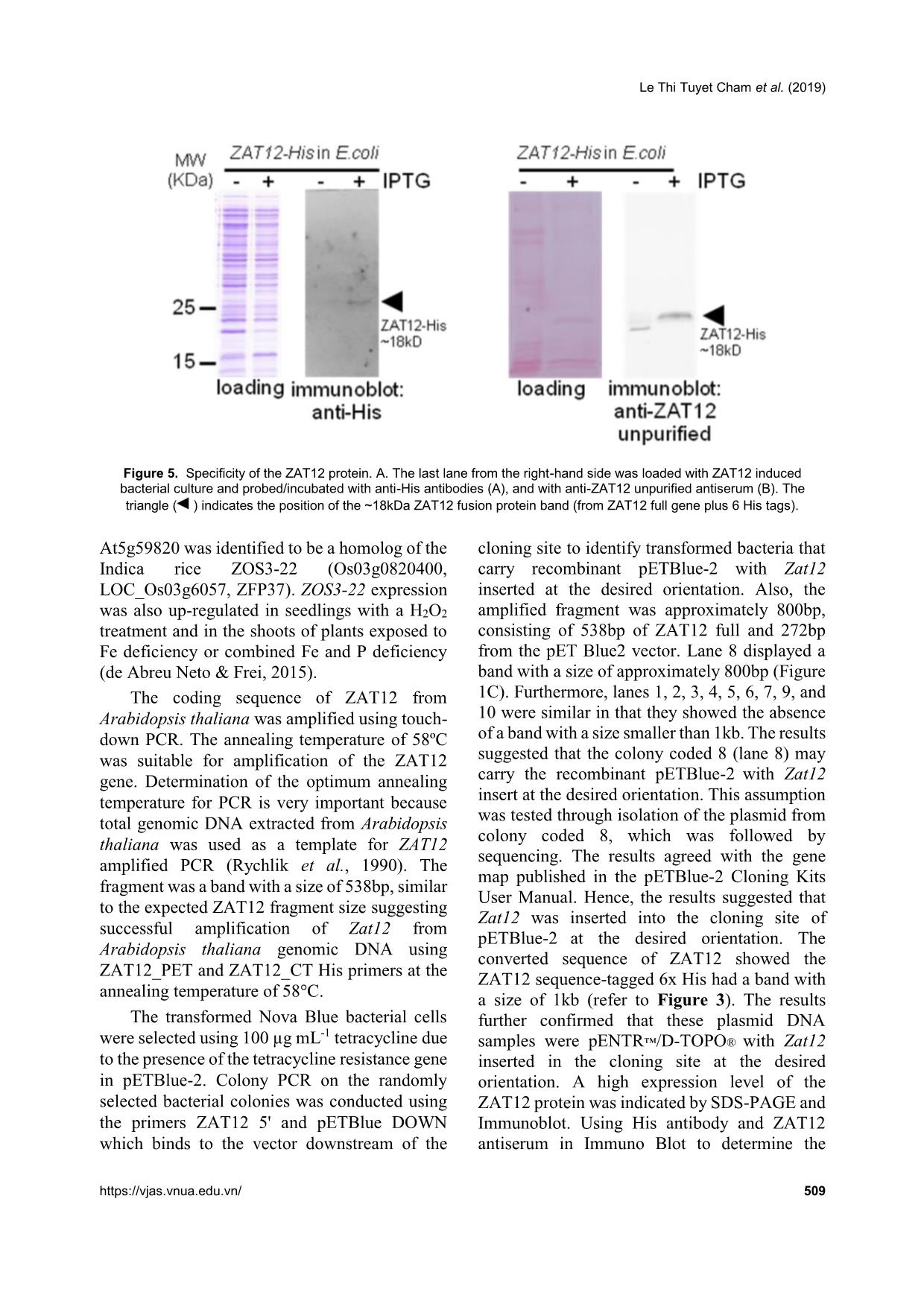
Trang 6

Trang 7

Trang 8
Tóm tắt nội dung tài liệu: Expression of Zinc Finger Protein Zat12 from Arabidopsis thaliana in Escherichia coli

, 0.5 M NaCl). The eluted antibody fractions were immediately neutralized by adding 1/10 volume of neutralization buffer (1 M Tris-HCl pH 8.0, 1.5 M NaCl, 1 mM EDTA, 0.5% NaN3) and BSA was added at a 1 mg mL-1 final concentration (Novagen, according to manufacturer’s manual). For the detection of the ZAT12-His protein, His antibodies and freshly purified undiluted anti- ZAT12 mouse antiserum were applied. These primary antibodies were detected with anti-mouse IgG conjugated with horseradish peroxidase (1:8000 dilution, Sigma-Aldrich, USA). The ZAT12-His protein was detected by incubation with His rat antibody (1:1000 Roche, Germany) and a secondary antibody anti-rat IgG (whole molecule)-horse radish peroxidase conjugate (1:10000, Sigma-Aldrich, USA). Detection signals were developed using an ZAT12 full DNA 5’ ZAT12_PET pETBlueDOWN 770 bp pETBlue-2 vector Le Thi Tuyet Cham et al. (2019) https://vjas.vnua.edu.vn/ 507 enhanced chemiluminescence detection kit (Biorad, USA) according to the manufacturer’s protocol. ZAT12 antibody preparation Based on the predicted antigenic propensity scores, a peptide corresponding to the N-terminal of ZAT12 was chemically synthesized and conjugated with KLH (Bio Trend) and later injected into mice to obtain a polyclonal antiserum (this work was conducted by Prof. U. Müller, Zoology Department, Saarland University). In this study, the antiserum was tested for its specificity by detecting bacteria positively expressing ZAT12. Results Plasmid construction and confirmation of cloned recombinant ZAT12 plasmid To generate the recombinant plasmid, the full-length DNA sequence of ZAT12 was specifically amplified with the ZAT12-PET and ZAT12-CT-His primers, which produced an expected band of 538bp. Upon successful ligation and transformation, the obtained colonies were numbered and a colony PCR was performed to check the positive clones for the presence of the recombinant plasmid. In addition, sequencing of the selected recombinant plasmid was performed to confirm the proper orientation of the insert by ligation (Figure 3). Resultant colonies were tested for the presence of the recombinant plasmid by colony PCR, and colonies were numbered as 1, 2, 3,, 10. If the insert was in the correct orientation, the expected size of the PCR product for ZAT12 with the primer combination (ZAT12 5' and pETBlueDOWN, see Figure 2) was approximately 800bp (538bp of ZAT12 full plus 232bp from the pET Blue2 vector). Colony no. 8 of ZAT12 gave a PCR product at the expected size. Expression of ZAT12 protein After verifying the sequence, the plasmid was transformed into BL21 (DE3) cells of E. coli for expression. SDS-PAGE analysis detected the expected products from BL21 (DE3) cells induced with IPTG. The results are shown in Figure 4. After colony PCR confirmation, the selected recombinant plasmid was transformed into the Tuner™ DE3 expression cells. Upon successful expression of the recombinant 18kDa ZAT12- His fusion protein at a small scale level, a large scale expression of ZAT12 protein was performed. Figure 3. Preparation of the ZAT12 encoding gene. A. The DNA fragment of ZAT12-His using PCR. B. Amino acid changes at the 5’-ends of ZAT12 introduced via PCR. C. Colony-PCR of ZAT12-His colonies was performed on 10 colonies. Resultant colonies were numbered as 1, 2, 3, ,10. Asterisks (*) indicate the ~800bp size ZAT12 fusion DNA band (from 538bp of ZAT12 full plus 232bp from the pET Blue2 vector). The only colony no. 8 gave a PCR product at the expected size, Figure 1A. M=ladder. Colony no. 8 was a positive colony. Expression of Zinc Finger Protein Zat12 from Arabidopsis thaliana in Escherichia coli 508 Vietnam Journal of Agricultural Sciences Figure 4. SDS-PAGE analysis of the heterologously expressed recombinant ZAT12 fusion protein in E. coli. ZAT12 fusion protein expression in Tuner™ DE3 cells accumulated as insoluble proteins (inclusion bodies) and soluble fraction. Asterisks (*) indicate the position of ~18kDa ZAT12 fusion protein band (from ZAT12 full gene plus 6 His tags). Specificity test of the ZAT12 protein The heterologously expressed recombinant ZAT12 was purified for affinity and injected into mice. The immunization of mice and collection of antiserum were kindly performed by Prof. Uli Müller and Iris Fuchs, Department of Zoology, Saarland University. The obtained antiserum was checked for its specificity on E. coli expressing the ZAT12 fusion protein (Figure 5). We detected a single band on western blot that matched to the specifically expressed and desired ZAT12 protein so that we were able to conclude that the ZAT12 protein was expressed successfully in E. coli. Discussion The C2H2-type plant-specific zinc finger transcription factor family was defined by the presence of a conserved zinc finger domain, in which two cysteines and two histidines coordinate with a single zinc atom to form a finger construct consisting of a short beta-hairpin and an alpha helix. At5g59820 was identified among the genes belonging to this family and named ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12). Structural analysis of the ZAT12 protein showed that it constituted of 162 amino acids divided into two C2H2-type zinc finger domains with a 22-amino acid inter- finger region, and a LDLSL core sequence of the EAR motif localized at the C terminus from amino acid 143 (Meissner & Michael, 1997; Englbrecht et al., 2004; Kagale et al., 2010). In plants, the ethylene-responsive element binding factor associated with the Amphiphilic Repression (EAR) motif is a transcriptional regulatory motif found as an active repressor in members of the ERF, C2H2, and AUX/IAA families, among others (Kagale et al., 2010). kDa Soluble fraction Insoluble fraction * * Le Thi Tuyet Cham et al. (2019) https://vjas.vnua.edu.vn/ 509 Figure 5. Specificity of the ZAT12 protein. A. The last lane from the right-hand side was loaded with ZAT12 induced bacterial culture and probed/incubated with anti-His antibodies (A), and with anti-ZAT12 unpurified antiserum (B). The triangle ( ) indicates the position of the ~18kDa ZAT12 fusion protein band (from ZAT12 full gene plus 6 His tags). At5g59820 was identified to be a homolog of the Indica rice ZOS3-22 (Os03g0820400, LOC_Os03g6057, ZFP37). ZOS3-22 expression was also up-regulated in seedlings with a H2O2 treatment and in the shoots of plants exposed to Fe deficiency or combined Fe and P deficiency (de Abreu Neto & Frei, 2015). The coding sequence of ZAT12 from Arabidopsis thaliana was amplified using touch- down PCR. The annealing temperature of 58ºC was suitable for amplification of the ZAT12 gene. Determination of the optimum annealing temperature for PCR is very important because total genomic DNA extracted from Arabidopsis thaliana was used as a template for ZAT12 amplified PCR (Rychlik et al., 1990). The fragment was a band with a size of 538bp, similar to the expected ZAT12 fragment size suggesting successful amplification of Zat12 from Arabidopsis thaliana genomic DNA using ZAT12_PET and ZAT12_CT His primers at the annealing temperature of 58°C. The transformed Nova Blue bacterial cells were selected using 100 µg mL-1 tetracycline due to the presence of the tetracycline resistance gene in pETBlue-2. Colony PCR on the randomly selected bacterial colonies was conducted using the primers ZAT12 5' and pETBlue DOWN which binds to the vector downstream of the cloning site to identify transformed bacteria that carry recombinant pETBlue-2 with Zat12 inserted at the desired orientation. Also, the amplified fragment was approximately 800bp, consisting of 538bp of ZAT12 full and 272bp from the pET Blue2 vector. Lane 8 displayed a band with a size of approximately 800bp (Figure 1C). Furthermore, lanes 1, 2, 3, 4, 5, 6, 7, 9, and 10 were similar in that they showed the absence of a band with a size smaller than 1kb. The results suggested that the colony coded 8 (lane 8) may carry the recombinant pETBlue-2 with Zat12 insert at the desired orientation. This assumption was tested through isolation of the plasmid from colony coded 8, which was followed by sequencing. The results agreed with the gene map published in the pETBlue-2 Cloning Kits User Manual. Hence, the results suggested that Zat12 was inserted into the cloning site of pETBlue-2 at the desired orientation. The converted sequence of ZAT12 showed the ZAT12 sequence-tagged 6x His had a band with a size of 1kb (refer to Figure 3). The results further confirmed that these plasmid DNA samples were pENTR™/D-TOPO® with Zat12 inserted in the cloning site at the desired orientation. A high expression level of the ZAT12 protein was indicated by SDS-PAGE and Immunoblot. Using His antibody and ZAT12 antiserum in Immuno Blot to determine the Expression of Zinc Finger Protein Zat12 from Arabidopsis thaliana in Escherichia coli 510 Vietnam Journal of Agricultural Sciences specificity of ZAT12 showed that ZAT12 was expressed successfully in E. coli. Conclusions Recombinant ZAT12 was expressed successfully in E.coli. This study helped accumulate enough ZAT12 recombinant protein for immunizing/injecting into mice in the following study of ZAT12 antibody generation. Acknowledgements The author would like to convey his her deep thanks to the Laboratory of Botany, Faculty of Biosciences, University, Saarbrueken, Germany for their kind hospitality and support in conducting this study. A special thanks to Professor Dr. Uli Mueller, Department of Zoology, Saarland University, for his kind help in producing anti-ZAT12 antiserum. References Ben Daniel B. H., Cattan E., Wachtel C., Avrahami D., Glick Y., Malichy A., Gerber D. & Miller G. (2016). Plant Identification of novel transcriptional regulators of Zat12 using comprehensive yeast one-hybrid screens. Physiologia Plantarum. 157(4): 422-441. Cheong Y. H., Chang H. S., Gupta R., Wang X., Zhu T. & Luan S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiology. 129(2): 661-677. Ciftci-Yilmaz S. & Mittler R. (2008). The zinc finger network of plants. Cellular and Molecular Life Sciences. 65: 1150-1160. Davletova S., Schlauch K., Coutu J. & Mittler R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology. 139(2): 847-856. De Abreu Neto J. B. & Frei M. (2015). Microarray meta- analysis focused on the response of genes involved in Redox Homeostasis to Diverse Abiotic stresses in rice. Frontiers in Plant Science. 6: 1260. Englbrecht C. C., Schoof H. & Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 5(1): 39. Fowler S. & Thomashow M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 14(8): 1675-1690. Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M. & Mackay J. P. (2007). Sticky fingers: zinc-fingers as protein-recognition motifs. Trends in Biochemical. Sciences. 32(2): 63-70. Iida A., Kazuoka T., Torikai S., Kikuchi H. & Oeda K. (2000). A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. The Plant Journal. 24(2): 191-203. Kagale S., Links M. G. & Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiology. 152(3): 1109-1134. Kiełbowicz-Matuk A. (2012). Involvement of plant C(2)H(2)-type zinc finger transcription factors in stress responses. Plant Science. 185-186: 78-85. Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X. & Harper J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 130(4): 2129-2141. Laemmli U. K. (1970). Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature. 227: 680-685. Le C. T. T., Brumbarova T., Ivanov R., Stoof C., Weber E., Mohrbacher J., Fink-Straube C. & Bauer P. (2016). Zinc finger of Arabidopsis thaliana 12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY- INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiology. 170(1): 540-557. Miller G., Shulaev V. & Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiologia Plantarum. 133(3): 481-489. Meissner R. & Michael A. J. (1997). Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Molecular Biology. 33(4): 615-624. Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci- Yilmaz S., Lee H., Stevenson B. & Zhu J. K. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Letters. 580(28-29): 6537-6542. Peng J., Li Z., Wen X., Li W., Shi H., Yang L., Zhu H. & Guo H. (2014). Salt induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genetics. 10(10): e1004664. Rizhsky L., Davletova S., Liang H. & Mittler R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. Journal of Biological Chemistry. 279(12): 11736-11743. Rychlik W., Spencer W. J. & Rhoads R. E. (1990). Optimization of the annealing temperature for amplification in vitro. Nucleic Acids Research. 18(21): 6409-6412. Le Thi Tuyet Cham et al. (2019) https://vjas.vnua.edu.vn/ 511 Sambrook J. & Rusell D. W. (2001). Molecular Cloning: A laboratory manual. Cold Spring Harbor Laboratory Press. 1(2-3):1-2344. Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inzé D. & Van Breusegem F. (2005). Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology. 139(2): 806-821. Vogel J. T., Zarka D. G., Van Buskirk H. A., Fowler S. G. & Thomashow M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal. 41(2): 195-211.
File đính kèm:
 expression_of_zinc_finger_protein_zat12_from_arabidopsis_tha.pdf
expression_of_zinc_finger_protein_zat12_from_arabidopsis_tha.pdf

