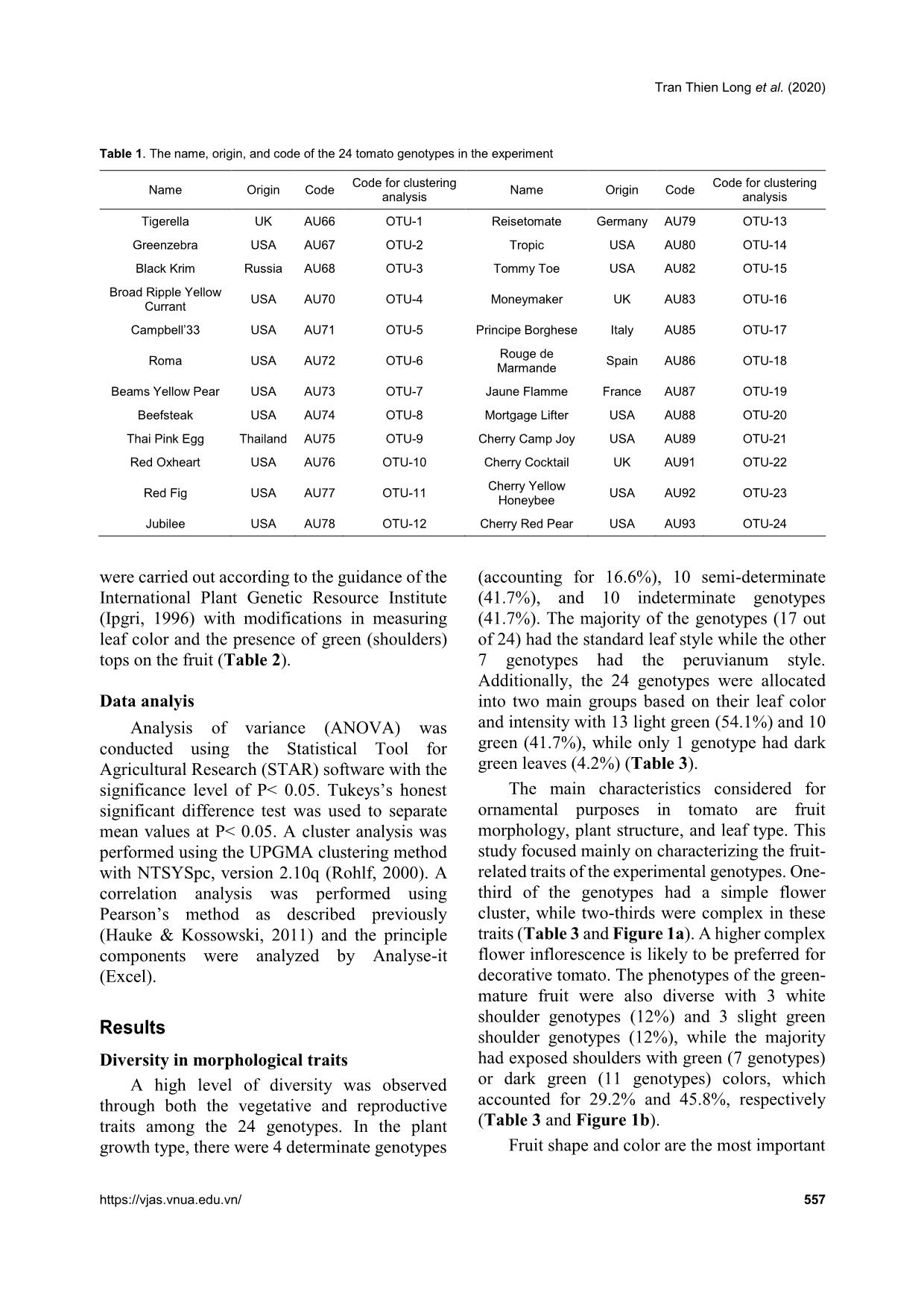
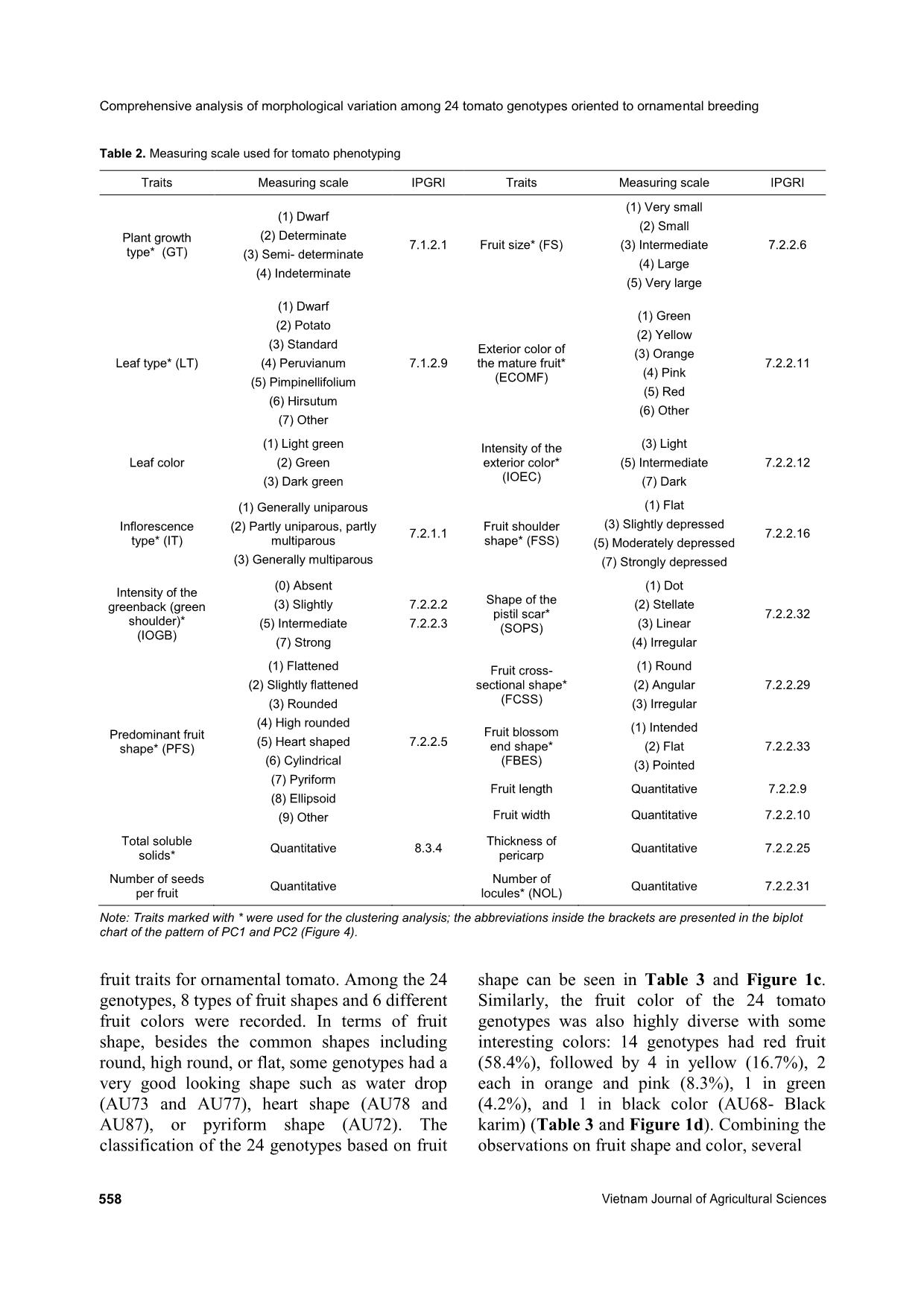
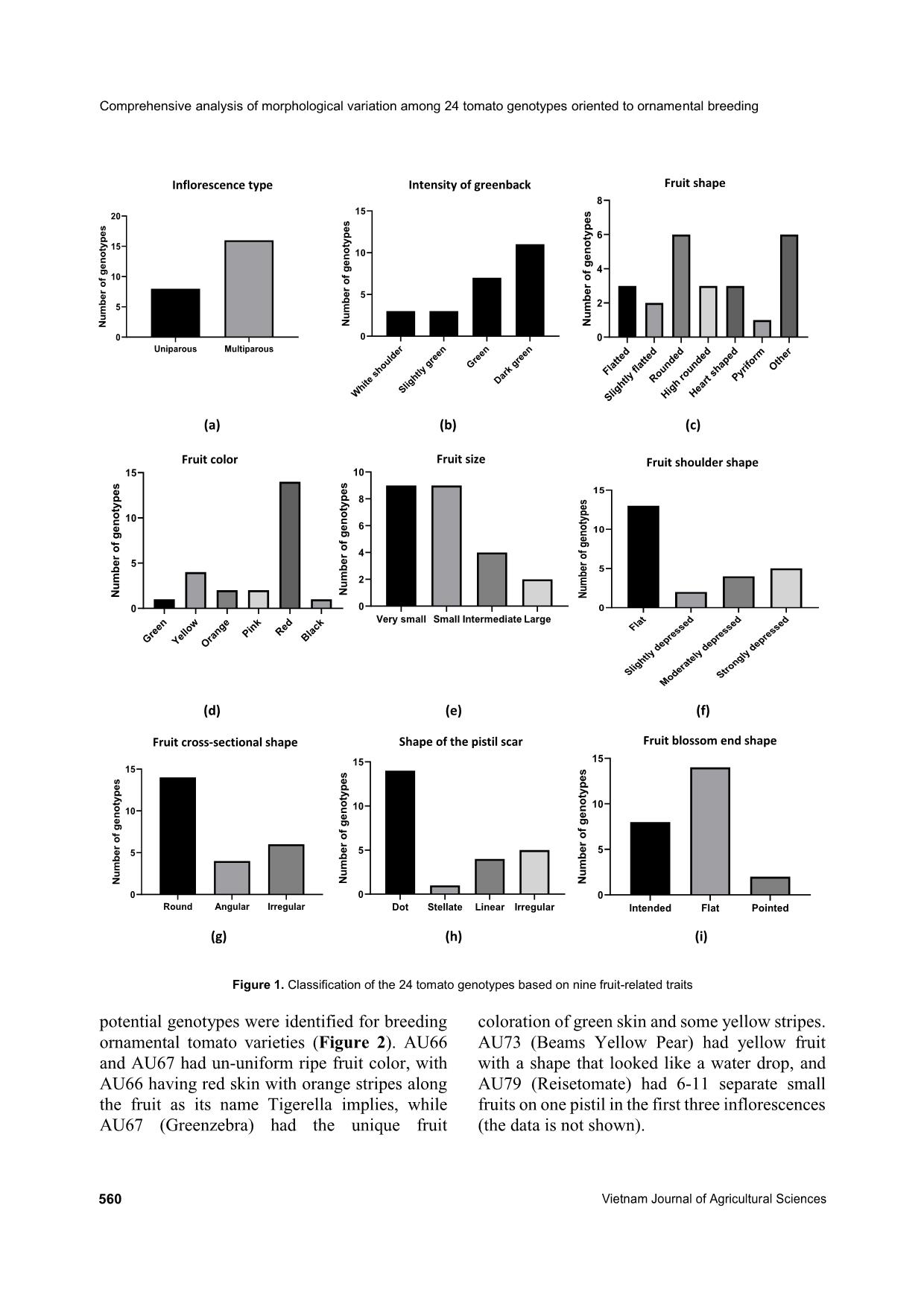
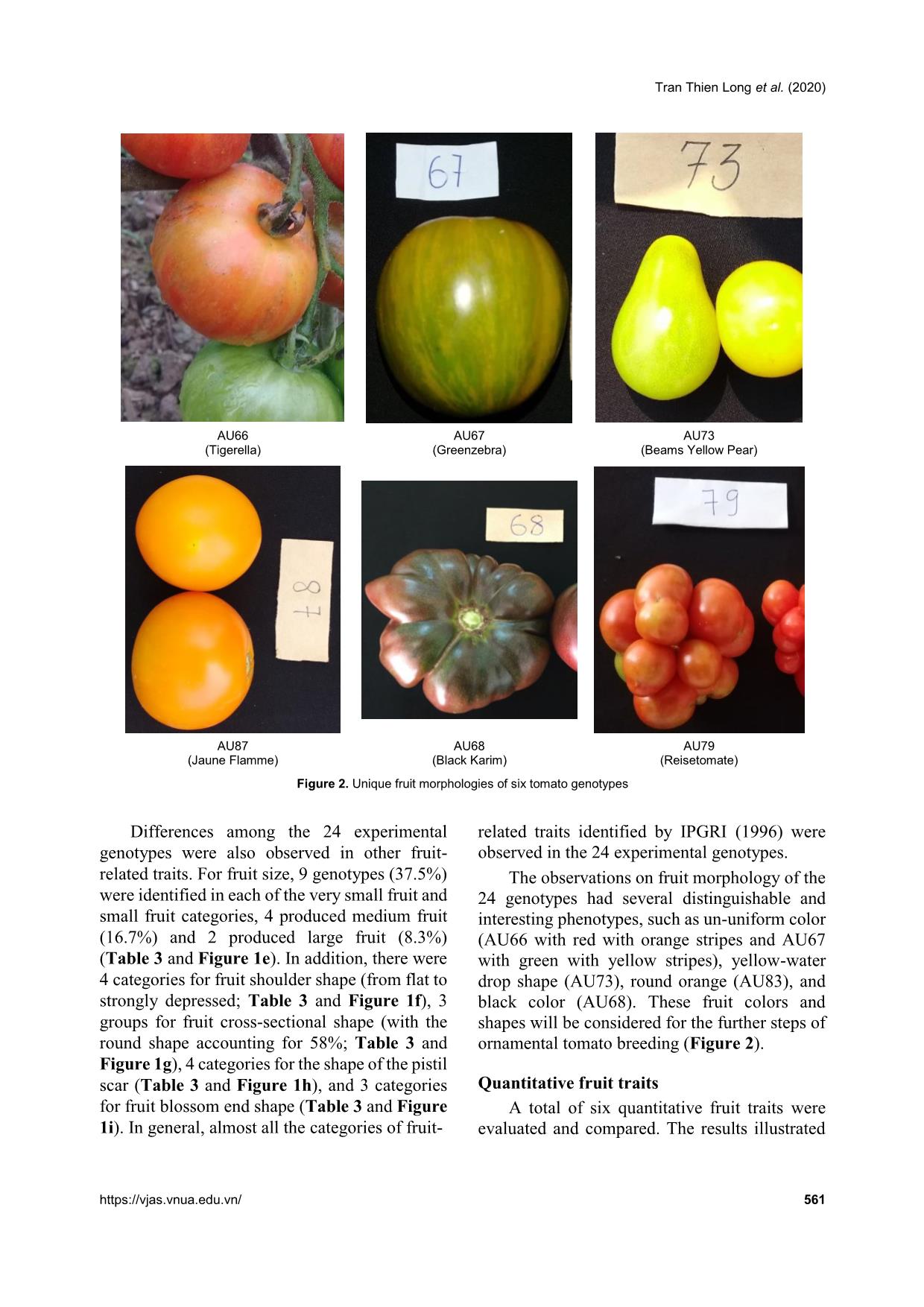
Comprehensive analysis of morphological variation among 24 tomato (Solanum lycopersicum) genotypes oriented to ornamental breeding in Vietnam
Tomato is one of the most important vegetables cultivated in
Vietnam. Besides its regular consumption as a vegetable, a new
demand for using tomato as a decorative plant on special occasions
was identified in recent years. This study aimed to characterize new
tomato accessions on their desirable morphological traits to select
potential materials for further breeding programs of ornamental
tomato varieties in Vietnam. Twenty-four heirloom tomato
genotypes were evaluated on 19 morphological traits. Based on the
describing system for tomato developed by the International Plant
Genetic Resources Institute (IPGRI, 1996), significant variation was
assessed in both qualitative and quantitative traits related to fruit
morphology. The results of principle component analysis indicated
that three main principle components explained over 60% of the total
phenotypic variation. The five traits of fruit size, fruit shoulder shape,
fruit cross-sectional shape, number of locules, and shape of the pistil
scar were recommended as important traits for clustering tomato
genotypes in this study. In addition, the 24 genotypes were classified
at the coefficient of 0.39 into six different clusters. Finally, six
interesting accessions, AU66, AU67, AU68, AU73, AU79, and
AU83 (with strange fruit colors and shapes), were selected as
potential materials for further breeding programs of ornamental
tomato in Vietnam.

Trang 1

Trang 2
Trang 3
Trang 4

Trang 5
Trang 6
Trang 7
Trang 8
Trang 9

Trang 10
Tải về để xem bản đầy đủ
Tóm tắt nội dung tài liệu: Comprehensive analysis of morphological variation among 24 tomato (Solanum lycopersicum) genotypes oriented to ornamental breeding in Vietnam

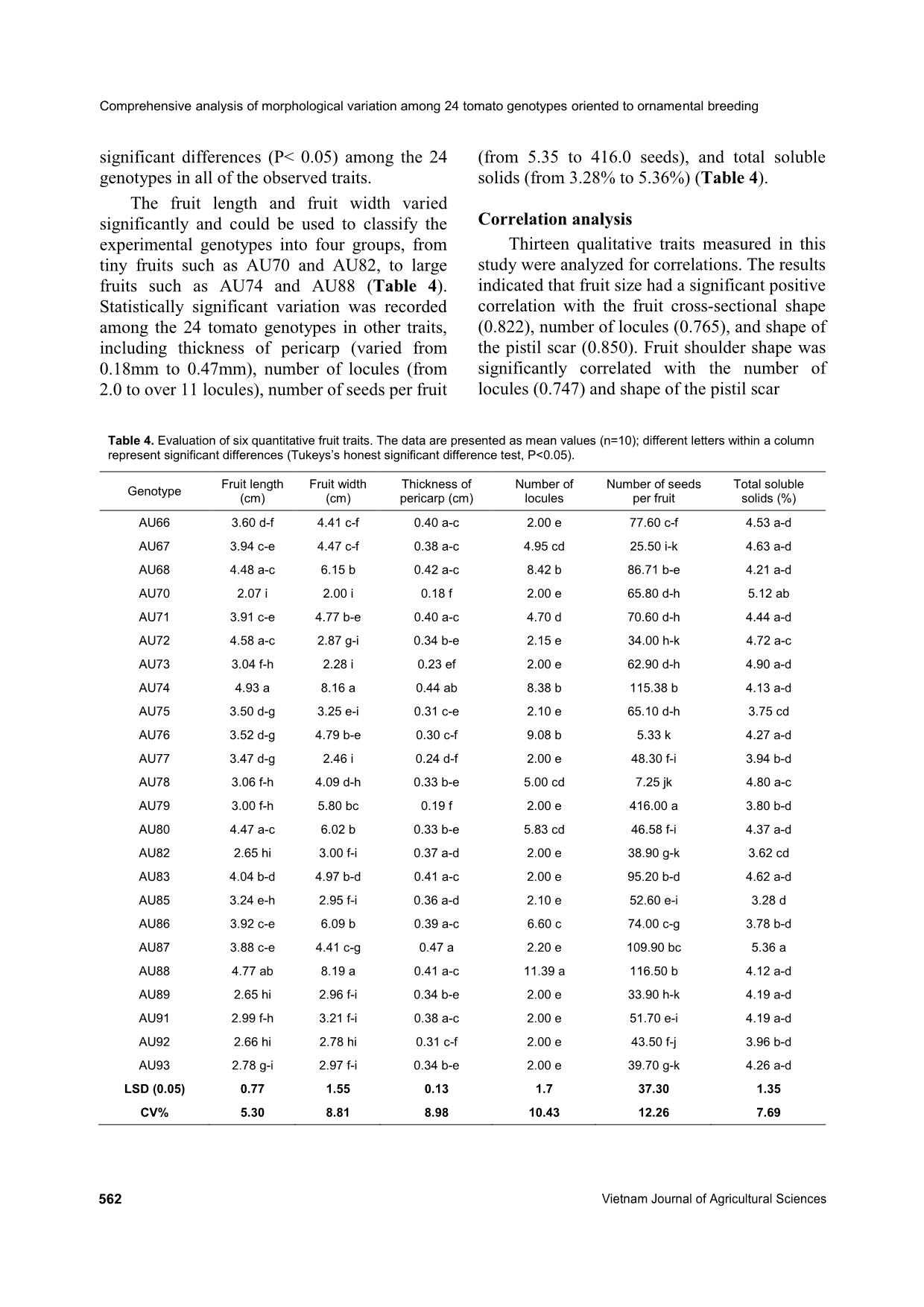
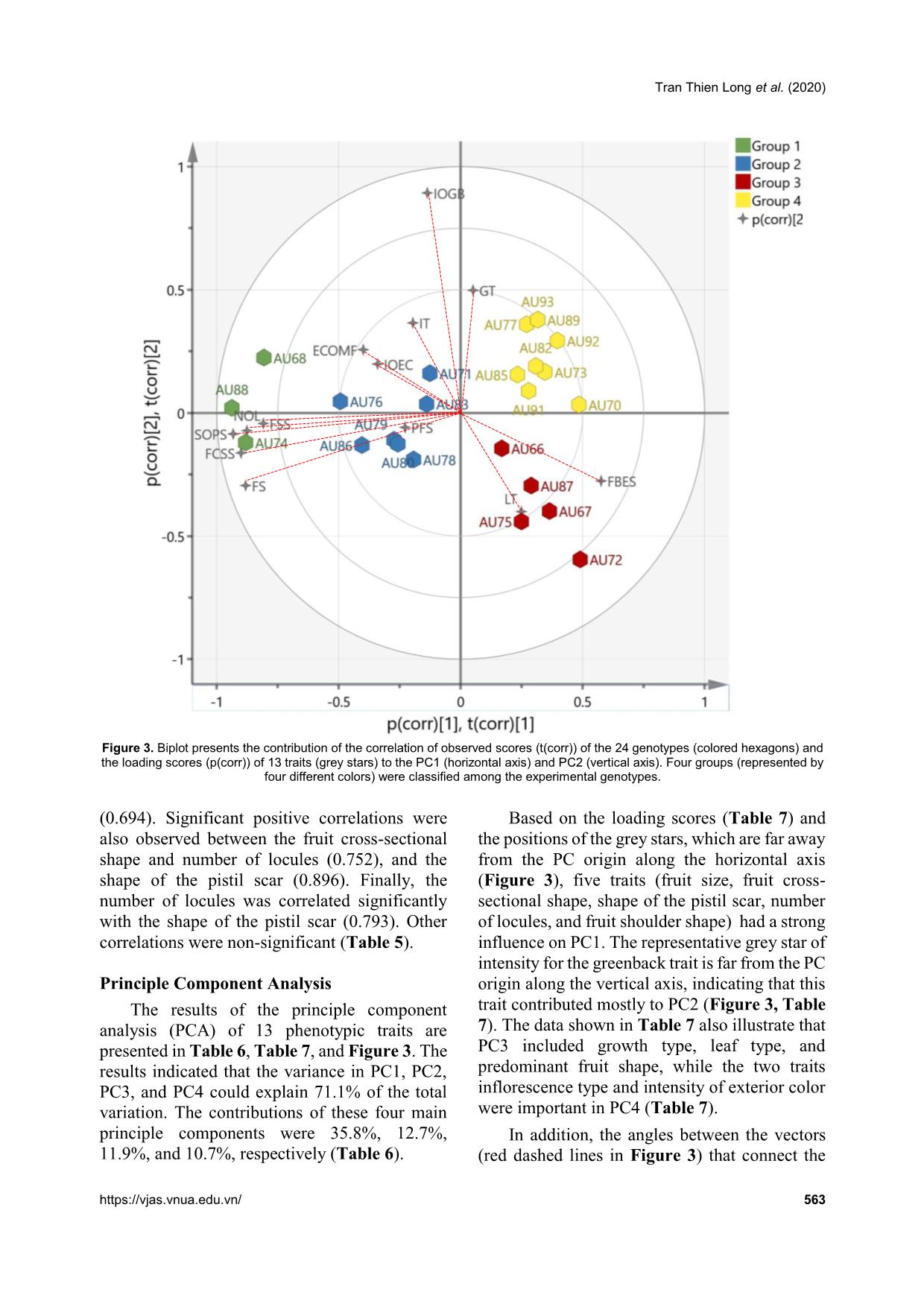
012; Cebolla-Cornejo et al., 2013). Conversely, the results of this present study suggested that the shape of the pistil scar should be considered as Tran Thien Long et al. (2020) https://vjas.vnua.edu.vn/ 565 Table 5. Pearson correlation coefficients between 13 traits of the 24 tomato genotypes Pearson's r Growth type Leaf type Inflorescence type Intensity of the greenback (green shoulder) Pre- dominant fruit shape Fruit size Exterior color of the mature fruit Intensity of the exterior color Fruit shoulder shape Fruit cross- sectional shape Number of locules Shape of the pistil scar Fruit blossom end shape Growth type 1.000 Leaf type 0.159 1.000 Inflorescence type 0.000 -0.324 1.000 Intensity of the greenback (green shoulder) 0.365 -0.247 0.202 1.000 Predominant fruit shape 0.142 0.204 0.165 -0.029 1.000 Fruit size -0.108 -0.168 -0.032 -0.171 0.135 1.000 Exterior color of the mature fruit -0.084 -0.211 0.150 0.180 0.089 0.233 1.000 Intensity of the exterior color 0.000 -0.225 -0.217 0.218 -0.107 0.328 0.149 1.000 Fruit shoulder shape -0.105 -0.243 0.237 0.028 0.110 0.612 0.314 0.247 1.000 Fruit cross-sectional shape -0.068 -0.072 0.243 0.007 0.298 0.822* 0.333 0.120 0.645 1.000 Number of locules 0.052 -0.111 0.079 0.105 0.176 0.765* 0.175 0.195 0.747* 0.752* 1.000 Shape of the pistil scar -0.092 -0.219 0.140 0.099 0.128 0.850* 0.265 0.324 0.694* 0.896* 0.793* 1.000 Fruit blossom end shape -0.146 -0.039 0.000 -0.382 -0.196 -0.468 -0.204 -0.171 -0.324 -0.412 -0.484 -0.445 1.000 Note: * is significant at the < 0.05 probability level. Comprehensive analysis of morphological variation among 24 tomato genotypes oriented to ornamental breeding 566 Vietnam Journal of Agricultural Sciences Table 6. The contributions of the principle components to variation among the 24 experimental genotypes based on 13 qualitative traits Component Variance Proportion Cumulative proportion 1 4.657 0.358 0.358 2 1.652 0.127 0.485 3 1.546 0.119 0.604 4 1.393 0.107 0.711 5 0.890 0.068 0.780 6 0.713 0.055 0.835 7 0.632 0.049 0.883 8 0.472 0.036 0.920 9 0.423 0.033 0.952 10 0.357 0.027 0.980 11 0.154 0.012 0.992 12 0.063 0.005 0.996 13 0.047 0.004 1.000 Table 7. Contributions of each qualitative trait to the main principle components Traits PC1 PC2 PC3 PC4 Growth type 0.051 0.497 0.598 0.125 Leaf type 0.248 -0.400 0.706 0.108 Inflorescence type -0.195 0.365 -0.174 -0.778 Intensity of the greenback (green shoulder) -0.138 0.894 0.103 0.124 Predominant fruit shape -0.227 -0.058 0.575 -0.420 Fruit size -0.881 -0.295 -0.011 0.154 Exterior color of the mature fruit -0.399 0.256 -0.214 -0.151 Intensity of the exterior color -0.341 0.198 -0.256 0.658 Fruit shoulder shape -0.809 -0.044 -0.148 -0.097 Fruit cross-sectional shape -0.900 -0.162 0.103 -0.182 Number of locules -0.876 -0.073 0.138 0.050 Shape of the pistil scar -0.933 -0.084 -0.046 0.048 Fruit blossom end shape 0.575 -0.277 -0.393 -0.205 an important trait for the main PCs (Table 5), which has not been reported in any previous study. Another aim of this study was to select suitable materials for ornamental tomato breeding. The results show that some genotypes have interesting traits, such as rare color or strange shape, that can be used for ornamental breeding. Previously, many reports have identified the genetic mechanisms of how tomato fruit shape and color are regulated. For example, different fruit colors in tomato are controlled independently or in interaction(s) among a group of genetic elements. Red tomato fruit is the most common color in nature (wild type) as well as in commercialized varieties, while the other colors are created on the background of this red color with different mutation(s). For instance, yellow Tran Thien Long et al. (2020) https://vjas.vnua.edu.vn/ 567 Figure 4. Cluster analysis of the 24 tomato genotypes based on 14 phenotypic traits (The analysis was conducted in NTSYSpc, version 2.10q using the UPMGA clustering method. The first dashed red line crosses the coefficient value of 0.39 which separates the 24 genotypes into 6 clusters while the second line is to identify the highest similar genotypes: AU77 and AU93 with a coefficient value of 0.93) Table 8. Six clusters derived from clustering the 24 tomato genotypes by UPMGA method Cluster Frequency Typical characters Genotype(s) I 4 White/slight green shoulder, small-red fruit, and flat fruit blossom end AU66, AU67, AU78, AU83 II 11 Indeterminate growth type, very small- red-round fruit, dotted pistil scar shape AU70, AU72, AU73, AU77, AU82, AU85, AU87, AU89, AU91, AU92, AU93 III 1 Dark green and moderately depressed fruit shoulder AU71 IV 1 Determinate growth type, white fruit shoulder, pink color AU75 V 6 Medium to large fruit size, irregular fruit shape and pistil scar, many locules per fruit AU68, AU74, AU79, AU80, AU86, AU88 VI 1 Dark green fruit shoulder, orange color, many locules AU76 is a recessive mutation on the R locus while while pink, orange, and green colors are controlled by mutation(s) on the Y locus, B and Del loci, and Gf locus, respectively (Liu et al., 2003). Conversely, black color is not naturally present in cultivated tomato but can be regulated by the genes Aft, atv, and abg from wild species (Jones et al., 2003; Canady et al., 2006; Mes et al., 2008). Similarly, tomato shape expression and control were also investigated comprehensively. Nine main shape categories were identified in tomato fruit (Visa et al., 2014) and four regions on chromosomes 2, 3, 7, and 8 carried the main loci related to the regulation of tomato fruit shape (Brewer et al., 2007). Overall, understanding the genetic regulation models of all colors and shapes in tomato enables researchers to use suitable breeding methods to create tomato materials with expected colors and shapes (for different ornamental purposes). In fact, some commercialized tomato cultivars were released for ornamental purposes by combining appropriate decorative traits, such as Sweet Valentine F1 (with a compact plant structure: 30- 40cm in height, spread 30-35cm; red heart- shaped fruit). The new fruit colors and shapes Comprehensive analysis of morphological variation among 24 tomato genotypes oriented to ornamental breeding 568 Vietnam Journal of Agricultural Sciences found in this study provide many ideas for ornamental breeding by combining interesting fruit morphologies with different plant structures (such as dwarf stem) and leaf types depending on the demands of customers. Conclusions The present study evaluated significant variation in 19 morphological characteristics including both qualitative and quantitative traits among 24 tomato genotypes. The 24 genotypes were also divided into 6 clusters based on the differences among 13 qualitative characteristics. The results of principle component analysis identified that three main PCs explained over 60% of the total phenotypic variation. In addition, six fruit traits (fruit size, fruit cross- sectional shape, fruit shoulder shape, number of locules, shape of the pistil scar, and intensity of the greenback) were recommended as important components of PC1 and PC2 in this study. Finally, six interesting accessions (with strange fruit colors and shapes) were identified as potential materials for further breeding programs of ornamental tomato (AU66, AU67, AU68, AU73, AU79, and AU87). Acknowledgements This work was supported by Vietnam National University of Agriculture under the Grant coded T2018-01-05. References AO C., Aiwansoba R. O., Osawaru M. E. & Ogwu M. C. (2017). Morphological evaluation of tomato (Solanum lycopersicum Linn.) Cultivars. Makara Journal of Science. 21(2): 97-106. Bhattarai K., Louws F. J., Williamson J. D. & Panthee D. R. (2016). Diversity analysis of tomato genotypes based on morphological traits with commercial breeding significance for fresh market production in eastern USA. Australian Journal of Crop Science. 10(8): 1098-1103. Bhattarai K., Sharma S. & Panthee D. R. (2018). Diversity among modern tomato genotypes at different levels in fresh-market breeding. International Journal of Agronomy. 2018: 1-15. Blanca J., Cañizares J., Cordero L., Pascual L., Diez M. J. & Nuez F. (2012). Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PloS One. 7(10): e48198. Brewer M. T., Moyseenko J. B., Monforte A. J. & van der Knaap E. (2007). Morphological variation in tomato: a comprehensive study of quantitative trait loci controlling fruit shape and development. Journal of Experimental Botany. 58(6): 1339-1349. Canady M. A., Ji Y. & Chetelat R. T. (2006). Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics. 174(4): 1775-1788. Cebolla-Cornejo J., Roselló S. & Nuez F. (2013). Phenotypic and genetic diversity of Spanish tomato landraces. Scientia Horticulturae. 162: 150-164. Danso Y., Akromah R. & Osei K. (2011). Molecular marker screening of tomato (Solanum lycopersicum L.) Germplasm for root-knot nematodes (Meloidogyne species) resistance. African Journal of Biotechnology. 10(9): 1511-1515. Glogovac S., Takač A., Tepić A., Šumić Z., Gvozdanović- Varga J., Červenski J., Vasić M. & Popović V. (2012). Principal component analysis of tomato genotypes based on some morphological and biochemical quality indicators. Ratarstvo i Povrtarstvo. 49(3): 296-301. Greene W. (2012). Vegetable Gardening the Colonial Williamsburg Way: 18th-Century Methods for Today's Organic Gardeners. In., Rodale Books: 240 pages. Ha T. M. (2015). Agronomic requirements and production methods of tomatoes in the Red River Delta of Vietnam. Journal of Tropical Crop Science. 2(1): 33- 38. Hanson P., Lu S.-F., Wang J.-F., Chen W., Kenyon L., Tan C.-W., Tee K. L., Wang Y.-Y., Hsu Y.-C. & Schafleitner R. (2016). Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Scientia Horticulturae. 201: 346-354. Hauke J. & Kossowski T. (2011). Comparison of values of Pearson's and Spearman's correlation coefficients on the same sets of data. Quaestiones Geographicae. 30(2): 87-93. Hoagland L., Navazio J., Zystro J., Kaplan I., Vargas J. G. & Gibson K. (2015). Key traits and promising germplasm for an organic participatory tomato breeding program in the US midwest. HortScience. 50(9): 1301-1308. Hu X., Wang H., Chen J. & Yang W. (2012). Genetic diversity of Argentina tomato varieties revealed by morphological traits, simple sequence repeat, and single nucleotide polymorphism markers. Pakistan Journal of Botany. 44(2): 485-492. IPGRI (1996). Descriptors for tomato (Lycopersicon spp.). In., Bioversity International: 47 pages. Jin L., Zhao L., Wang Y., Zhou R., Song L., Xu L., Cui X., Li R., Yu W. & Zhao T. (2019). Genetic diversity of Tran Thien Long et al. (2020) https://vjas.vnua.edu.vn/ 569 324 cultivated tomato germplasm resources using agronomic traits and InDel markers. Euphytica. 215(4): 69-84. Jones C., Mes P. & Myers J. (2003). Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. Journal of Heredity. 94(6): 449-456. Kaur S., Singh A., Bagati S., Sharma M. & Sharma S. (2019). Morphological Markers based Assessment of Genetic Diversity in Cultivated Tomato (Solanum Lycopersicon L.) Genotypes. International Journal of Environment, Agriculture and Biotechnology. 3(2): 567-573. Liu Y. S., Gur A., Ronen G., Causse M., Damidaux R., Buret M., Hirschberg J. & Zamir D. (2003). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnology Journal. 1(3): 195-207. Martí E., Gisbert C., Bishop G. J., Dixon M. S. & García- Martínez J. L. (2006). Genetic and physiological characterization of tomato cv. Micro-Tom. Journal of Experimental Botany. 57(9): 2037-2047. Mes P. J., Boches P., Myers J. R. & Durst R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. Journal of the American Society for Horticultural Science. 133(2): 262-269. Omondi S. (2017). The most popular vegetable in the world [Online]. Retrieved from https://www.worldatlas.com/articles/the-most- popular-vegetables-in-the-world.html on July 15 2020. Panthee D. R. & Chen F. (2010). Genomics of fungal disease resistance in tomato. Current Genomics. 11(1): 30-39. Panthee D. R., Labate J. A. & Robertson L. D. (2013). Evaluation of tomato accessions for flavour and flavour-contributing components. Plant Genetic Resources. 11(2): 106-113. Reddy B., Reddy M. P., Begum H. & Sunil N. (2013). Genetic diversity studies in tomato (Solanum lycopersicum L.). Journal of Agriculture and Veterinary Science. 4(4): 55-53. Rohlf F. J. (2000). NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.1 Exeter Software. Setauket, New York, USA. Saito T., Ariizumi T., Okabe Y., Asamizu E., Hiwasa- Tanase K., Fukuda N., Mizoguchi T., Yamazaki Y., Aoki K. & Ezura H. (2011). TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiology. 52(2): 283- 296. Singh M., Singh A., Kumar A. & Pandey K. (2018). Molecular diversity of tomato germplasm (Lycopersicum esculentum L.) using lycopene specific markers. Biocatalysis and Agricultural Biotechnology. 16: 340-346. Srinivasan R. (2010). Safer tomato production techniques: a field guide for soil fertility and pest management. In., AVRDC–The World Vegetable Center: 97 pages. Tembe K. O., Chemining'wa G., Ambuko J. & Owino W. (2018). Evaluation of African tomato landraces (Solanum lycopersicum) based on morphological and horticultural traits. Agriculture and Natural Resources. 52(6): 536-542. Visa S., Cao C., Gardener B. M. & van der Knaap E. (2014). Modeling of tomato fruits into nine shape categories using elliptic fourier shape modeling and Bayesian classification of contour morphometric data. Euphytica. 200(3): 429-439. Wang T., Zou Q., Qi S., Wang X., Wu Y., Liu N., Zhang Y., Zhang Z. & Li H. (2016). Analysis of genetic diversity and population structure in a tomato (Solanum lycopersicum L.) germplasm collection based on single nucleotide polymorphism markers. Genetics and Molecular Research. 15(3): 1-12. Ziaf K., Amjad M., Shakeel A., Azhar M. & Saeed A. (2016). Assessment of genetic diversity in tomato for fruit morphology, composition and yield. Pakistan Journal of Botany. 48(6): 2477-2483.
File đính kèm:
 comprehensive_analysis_of_morphological_variation_among_24_t.pdf
comprehensive_analysis_of_morphological_variation_among_24_t.pdf

